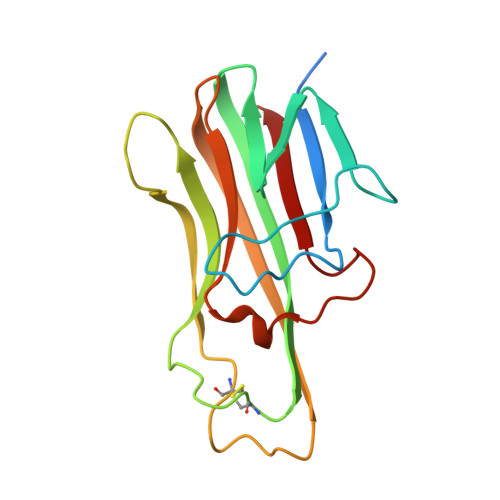
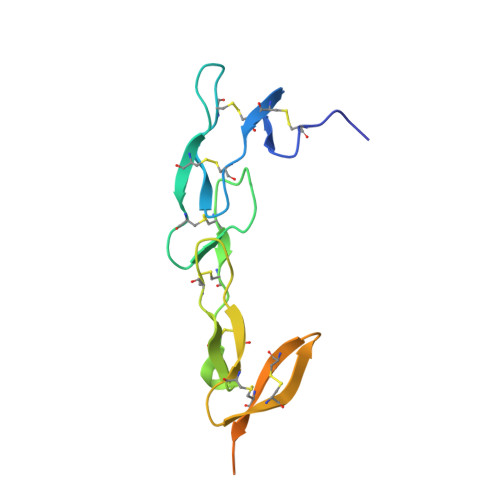
HVEM structures and mutants reveal distinct functions of binding to LIGHT and BTLA/CD160.
Liu, W., Chou, T.F., Garrett-Thomson, S.C., Seo, G.Y., Fedorov, E., Ramagopal, U.A., Bonanno, J.B., Wang, Q., Kim, K., Garforth, S.J., Kakugawa, K., Cheroutre, H., Kronenberg, M., Almo, S.C.(2021) J Exp Medicine 218
- PubMed: 34709351
- DOI: https://doi.org/10.1084/jem.20211112
- Primary Citation of Related Structures:
4RSU, 7MSG, 7MSJ - PubMed Abstract:
HVEM is a TNF (tumor necrosis factor) receptor contributing to a broad range of immune functions involving diverse cell types. It interacts with a TNF ligand, LIGHT, and immunoglobulin (Ig) superfamily members BTLA and CD160. Assessing the functional impact of HVEM binding to specific ligands in different settings has been complicated by the multiple interactions of HVEM and HVEM binding partners. To dissect the molecular basis for multiple functions, we determined crystal structures that reveal the distinct HVEM surfaces that engage LIGHT or BTLA/CD160, including the human HVEM-LIGHT-CD160 ternary complex, with HVEM interacting simultaneously with both binding partners. Based on these structures, we generated mouse HVEM mutants that selectively recognized either the TNF or Ig ligands in vitro. Knockin mice expressing these muteins maintain expression of all the proteins in the HVEM network, yet they demonstrate selective functions for LIGHT in the clearance of bacteria in the intestine and for the Ig ligands in the amelioration of liver inflammation.
- Department of Biochemistry, Albert Einstein College of Medicine, Bronx, NY.
Organizational Affiliation: