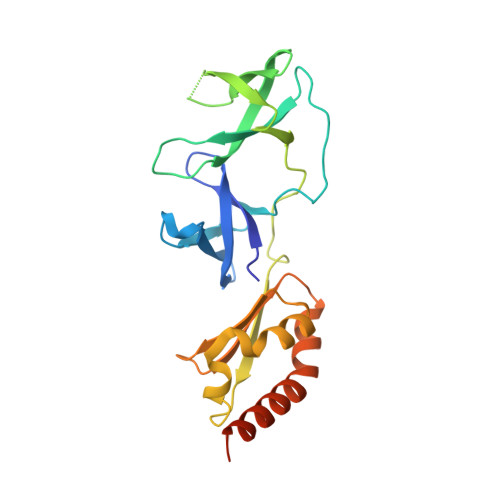
Human FMRP contains an integral tandem Agenet (Tudor) and KH motif in the amino terminal domain.
Myrick, L.K., Hashimoto, H., Cheng, X., Warren, S.T.(2015) Hum Mol Genet 24: 1733-1740
- PubMed: 25416280
- DOI: https://doi.org/10.1093/hmg/ddu586
- Primary Citation of Related Structures:
4QVZ, 4QW2 - PubMed Abstract:
Fragile X syndrome, a common cause of intellectual disability and autism, is due to mutational silencing of the FMR1 gene leading to the absence of its gene product, fragile X mental retardation protein (FMRP). FMRP is a selective RNA binding protein owing to two central K-homology domains and a C-terminal arginine-glycine-glycine (RGG) box. However, several properties of the FMRP amino terminus are unresolved. It has been documented for over a decade that the amino terminus has the ability to bind RNA despite having no recognizable functional motifs. Moreover, the amino terminus has recently been shown to bind chromatin and influence the DNA damage response as well as function in the presynaptic space, modulating action potential duration. We report here the amino terminal crystal structures of wild-type FMRP, and a mutant (R138Q) that disrupts the amino terminus function, containing an integral tandem Agenet and discover a novel KH motif.
- Department of Human Genetics.
Organizational Affiliation: