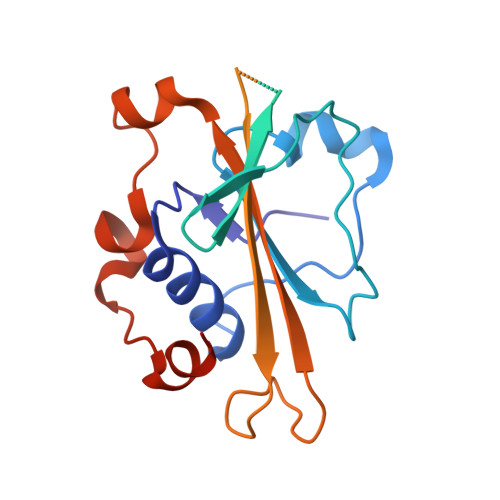
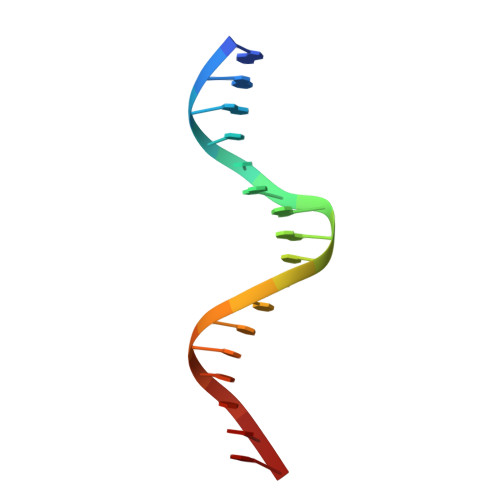
Crystal structure of the WOPR-DNA complex and implications for Wor1 function in white-opaque switching of Candida albicans.
Zhang, S., Zhang, T., Yan, M., Ding, J., Chen, J.(2014) Cell Res 24: 1108-1120
- PubMed: 25091450
- DOI: https://doi.org/10.1038/cr.2014.102
- Primary Citation of Related Structures:
4QTJ, 4QTK - PubMed Abstract:
Wor1 (white-opaque switching regulator 1) is a master regulator of the white-opaque switching in Candida albicans, an opportunistic human fungal pathogen, and is associated with its pathogenicity and commensality. Wor1 contains a conserved DNA-binding region at the N-terminus, consisting of two conserved segments (WOPRa and WOPRb) connected by a non-conserved linker that can bind to specific DNA sequences of the promoter regions and then regulates the transcription. Here, we report the crystal structure of the C. albicans Wor1 WOPR segments in complex with a double-stranded DNA corresponding to one promoter region of WOR1. The sequentially separated WOPRa and WOPRb are structurally interwound together to form a compact globular domain that we term the WOPR domain. The WOPR domain represents a new conserved fungal-specific DNA-binding domain which uses primarily a conserved loop to recognize and interact specifically with a conserved 6-bp motif of the DNA in both minor and major grooves. The protein-DNA interactions are essential for WOR1 transcriptional regulation and white-to-opaque switching. The structural and biological data together reveal the molecular basis for the recognition and binding specificity of the WOPR domain with its specific DNA sequences and the function of Wor1 in the activation of transcription.
- State Key Laboratory of Molecular Biology, Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, 320 Yue Yang Road, Shanghai 200031, China.
Organizational Affiliation: