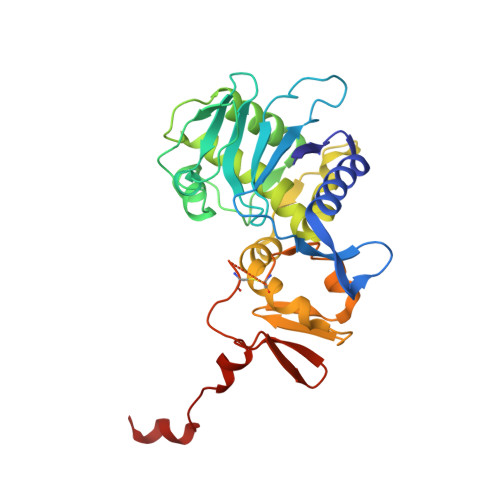
The Molecular Mechanism of Shiga Toxin Stx2e Neutralization by a Single-domain Antibody Targeting the Cell Receptor-binding Domain.
Lo, A.W., Moonens, K., De Kerpel, M., Brys, L., Pardon, E., Remaut, H., De Greve, H.(2014) J Biological Chem 289: 25374-25381
- PubMed: 25053417
- DOI: https://doi.org/10.1074/jbc.M114.566257
- Primary Citation of Related Structures:
4P2C - PubMed Abstract:
Shiga toxin Stx2e is the major known agent that causes edema disease in newly weaned pigs. This severe disease is characterized by neurological disorders, hemorrhagic lesions, and frequent fatal outcomes. Stx2e consists of an enzymatically active A subunit and five B subunits that bind to a specific glycolipid receptor on host cells. It is evident that antibodies binding to the A subunit or the B subunits of Shiga toxin variants may have the capability to inhibit their cytotoxicity. Here, we report the discovery and characterization of a VHH single domain antibody (nanobody) isolated from a llama phage display library that confers potent neutralizing capacity against Stx2e toxin. We further present the crystal structure of the complex formed between the nanobody (NbStx2e1) and the Stx2e toxoid, determined at 2.8 Å resolution. Structural analysis revealed that for each B subunit of Stx2e, one NbStx2e1 is interacting in a head-to-head orientation and directly competing with the glycolipid receptor binding site on the surface of the B subunit. The neutralizing NbStx2e1 can in the future be used to prevent or treat edema disease.
- From Structural and Molecular Microbiology, Structural Biology Research Center, VIB, Structural Biology Brussels, and.
Organizational Affiliation: