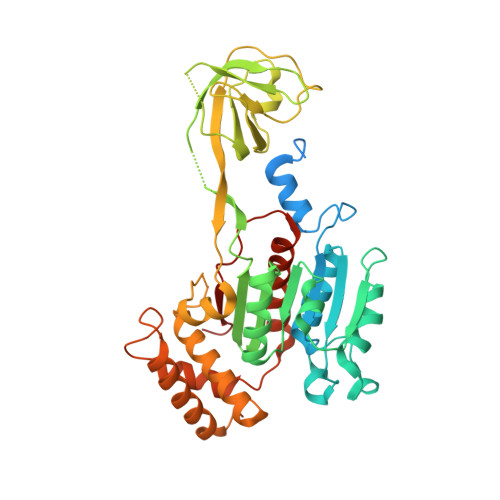
Expression, purification, and crystal structure of N-terminal domains of human ubiquitin-activating enzyme (E1).
Xie, S.T.(2014) Biosci Biotechnol Biochem 78: 1542-1549
- PubMed: 25209502
- DOI: https://doi.org/10.1080/09168451.2014.923301
- Primary Citation of Related Structures:
4P22 - PubMed Abstract:
Ubiquitin-activating enzyme (E1) is a key regulator in protein ubiquitination, which lies on the upstream of the ubiquitin-related pathways and determines the activation of the downstream enzyme cascade. Thus far, no structural information about the human ubiquitin-activating enzyme has been reported. We expressed and purified the N-terminal domains of human E1 and determined their crystal structures, which contain inactive adenylation domain (IAD) and the first catalytic cysteine half-domain (FCCH). This study presents the crystal structure of human E1 fragment for the first time. The main structure of both IAD and FCCH superimposed well with their corresponding domains in yeast Uba1, but their relative positions vary significantly. This work provides new structural insights in understanding the mechanisms of ubiquitin activation in humans.
- a MOE Key Laboratory of Protein Sciences and Tsinghua-Peking Center for Life Sciences , School of Life Sciences, Tsinghua University , Beijing , China.
Organizational Affiliation: