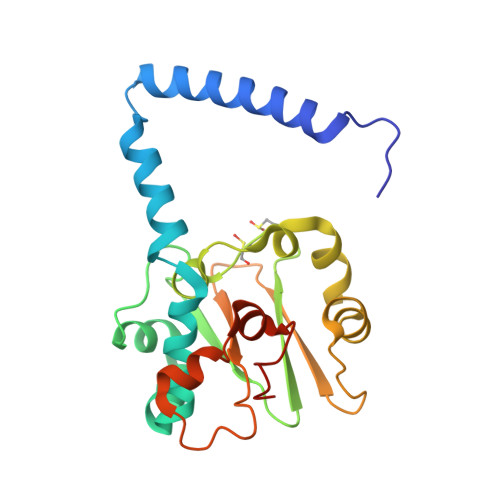
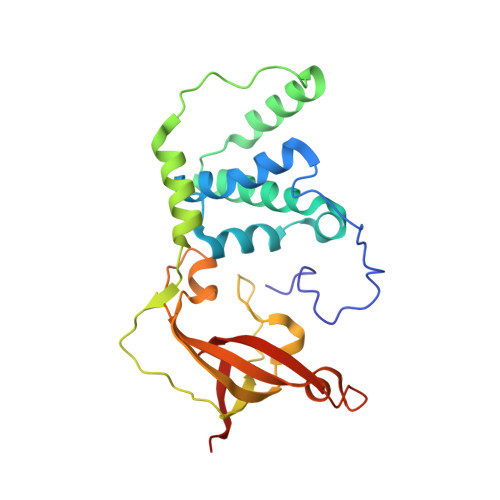
The active site sulfenic acid ligand in nitrile hydratases can function as a nucleophile.
Martinez, S., Wu, R., Sanishvili, R., Liu, D., Holz, R.(2014) J Am Chem Soc 136: 1186-1189
- PubMed: 24383915
- DOI: https://doi.org/10.1021/ja410462j
- Primary Citation of Related Structures:
4OB0, 4OB1, 4OB2, 4OB3 - PubMed Abstract:
Nitrile hydratase (NHase) catalyzes the hydration of nitriles to their corresponding commercially valuable amides at ambient temperatures and physiological pH. Several reaction mechanisms have been proposed for NHase enzymes; however, the source of the nucleophile remains a mystery. Boronic acids have been shown to be potent inhibitors of numerous hydrolytic enzymes due to the open shell of boron, which allows it to expand from a trigonal planar (sp(2)) form to a tetrahedral form (sp(3)). Therefore, we examined the inhibition of the Co-type NHase from Pseudonocardia thermophila JCM 3095 (PtNHase) by boronic acids via kinetics and X-ray crystallography. Both 1-butaneboronic acid (BuBA) and phenylboronic acid (PBA) function as potent competitive inhibitors of PtNHase. X-ray crystal structures for BuBA and PBA complexed to PtNHase were solved and refined at 1.5, 1.6, and 1.2 Å resolution. The resulting PtNHase-boronic acid complexes represent a "snapshot" of reaction intermediates and implicate the cysteine-sulfenic acid ligand as the catalytic nucleophile, a heretofore unknown role for the αCys(113)-OH sulfenic acid ligand. Based on these data, a new mechanism of action for the hydration of nitriles by NHase is presented.
- Department of Chemistry, Marquette University , Milwaukee, Wisconsin 53201, United States.
Organizational Affiliation: