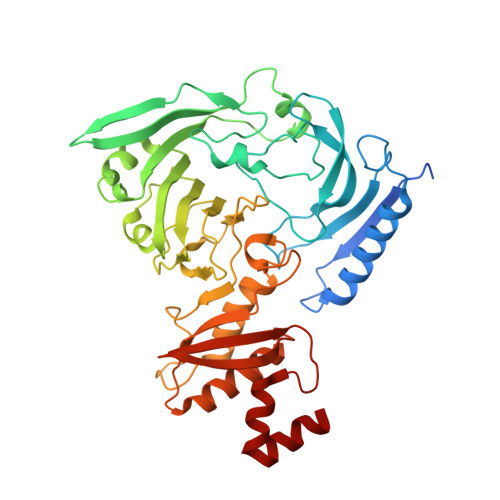
A structurally distinct human mycoplasma protein that generically blocks antigen-antibody union.
Grover, R.K., Zhu, X., Nieusma, T., Jones, T., Boero, I., MacLeod, A.S., Mark, A., Niessen, S., Kim, H.J., Kong, L., Assad-Garcia, N., Kwon, K., Chesi, M., Smider, V.V., Salomon, D.R., Jelinek, D.F., Kyle, R.A., Pyles, R.B., Glass, J.I., Ward, A.B., Wilson, I.A., Lerner, R.A.(2014) Science 343: 656-661
- PubMed: 24503852
- DOI: https://doi.org/10.1126/science.1246135
- Primary Citation of Related Structures:
4NZR, 4NZT, 4NZU - PubMed Abstract:
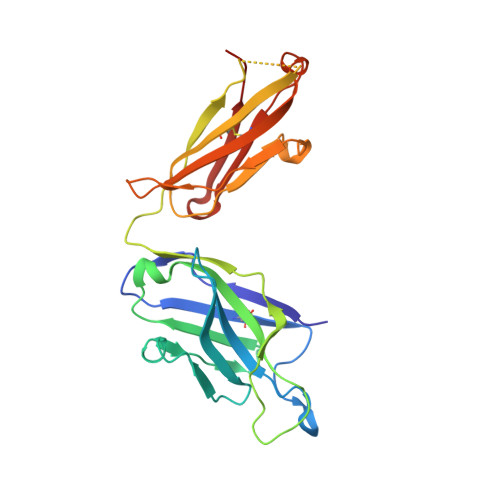
We report the discovery of a broadly reactive antibody-binding protein (Protein M) from human mycoplasma. The crystal structure of the ectodomain of transmembrane Protein M differs from other known protein structures, as does its mechanism of antibody binding. Protein M binds with high affinity to all types of human and nonhuman immunoglobulin G, predominantly through attachment to the conserved portions of the variable region of the κ and λ light chains. Protein M blocks antibody-antigen union, likely because of its large C-terminal domain extending over the antibody-combining site, blocking entry to large antigens. Similar to the other immunoglobulin-binding proteins such as Protein A, Protein M as well as its orthologs in other Mycoplasma species could become invaluable reagents in the antibody field.
- Department of Cell and Molecular Biology, The Scripps Research Institute, La Jolla, CA 92037, USA.
Organizational Affiliation: