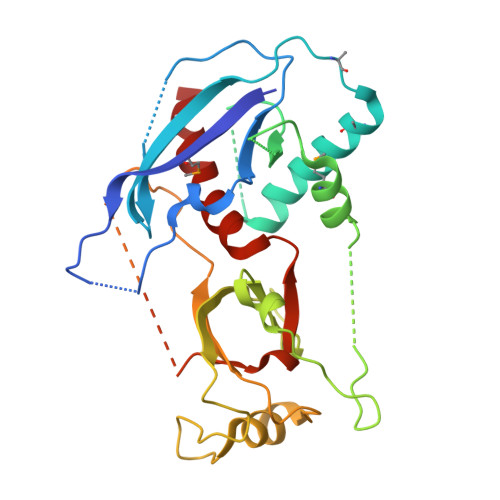
A Phosphate-Binding Pocket within the Platform-PAZ-Connector Helix Cassette of Human Dicer.
Tian, Y., Simanshu, D.K., Ma, J.B., Park, J.E., Heo, I., Kim, V.N., Patel, D.J.(2014) Mol Cell 53: 606-616
- PubMed: 24486018
- DOI: https://doi.org/10.1016/j.molcel.2014.01.003
- Primary Citation of Related Structures:
4NGB, 4NGC, 4NGD, 4NGF, 4NGG, 4NH3, 4NH5, 4NH6, 4NHA - PubMed Abstract:
We have solved two families of crystal structures of the human Dicer "platform-PAZ-connector helix" cassette in complex with small interfering RNAs (siRNAs). The structures possess two adjacently positioned pockets: a 2 nt 3'-overhang-binding pocket within the PAZ domain (3' pocket) and a phosphate-binding pocket within the platform domain (phosphate pocket). One family of complexes contains a knob-like α-helical protrusion, designated "hDicer-specific helix," that separates the two pockets and orients the bound siRNA away from the surface of Dicer, which could be indicative of a product release/transfer state. In the second complex, the helical protrusion is melted/disordered and the bound siRNA is aligned toward the surface of Dicer, suggestive of a cleavage-competent state. These structures allow us to propose that the transition from the cleavage-competent to the postulated product release/transfer state may involve release of the 5'-phosphate from the phosphate pocket while retaining the 3' overhang in the 3' pocket.
- Structural Biology Program, Memorial Sloan-Kettering Cancer Center, New York, NY 10065, USA.
Organizational Affiliation: