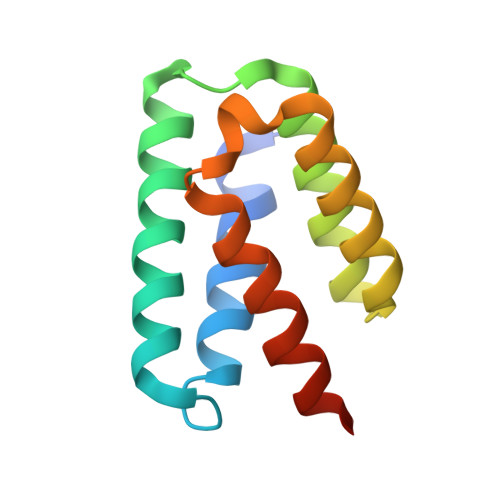
Structure and function of steroid receptor RNA activator protein, the proposed partner of SRA noncoding RNA.
McKay, D.B., Xi, L., Barthel, K.K., Cech, T.R.(2014) J Mol Biology 426: 1766-1785
- PubMed: 24486609
- DOI: https://doi.org/10.1016/j.jmb.2014.01.006
- Primary Citation of Related Structures:
4NBO - PubMed Abstract:
In a widely accepted model, the steroid receptor RNA activator protein (SRA protein; SRAP) modulates the transcriptional regulatory activity of SRA RNA by binding a specific stem-loop of SRA. We first confirmed that SRAP is present in the nucleus as well as the cytoplasm of MCF-7 breast cancer cells, where it is expressed at the level of about 10(5) molecules per cell. However, our SRAP-RNA binding experiments, both in vitro with recombinant protein and in cultured cells with plasmid-expressed protein and RNA, did not reveal a specific interaction between SRAP and SRA. We determined the crystal structure of the carboxy-terminal domain of human SRAP and found that it does not have the postulated RRM (RNA recognition motif). The structure is a five-helix bundle that is distinct from known RNA-binding motifs and instead is similar to the carboxy-terminal domain of the yeast spliceosome protein PRP18, which stabilizes specific protein-protein interactions within a multisubunit mRNA splicing complex. SRA binding experiments with this domain gave negative results. Transcriptional regulation by SRA/SRAP was examined with siRNA knockdown. Effects on both specific estrogen-responsive genes and genes identified by RNA-seq as candidates for regulation were examined in MCF-7 cells. Only a small effect (~20% change) on one gene resulting from depletion of SRA/SRAP could be confirmed. We conclude that the current model for SRAP function must be reevaluated; we suggest that SRAP may function in a different context to stabilize specific intermolecular interactions in the nucleus.
- BioFrontiers Institute, University of Colorado, Boulder, CO 80309, USA.
Organizational Affiliation: