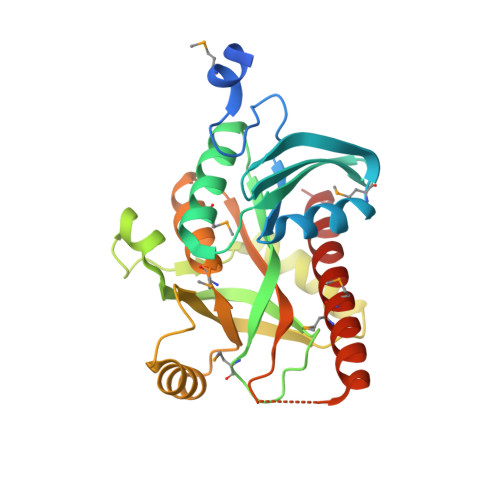
Crystal structure of uridine phosphorylase from vibrio fischeri es114 complexed with 6-hydroxy-1-naphthoic acid, NYSGRC Target 029520.
Malashkevich, V.N., Bhosle, R., Toro, R., Hillerich, B., Gizzi, A., Garforth, S., Kar, A., Chan, M.K., Lafluer, J., Patel, H., Matikainen, B., Chamala, S., Lim, S., Celikgil, A., Villegas, G., Evans, B., Love, J., Fiser, A., Khafizov, K., Seidel, R., Bonanno, J.B., Almo, S.C.To be published.