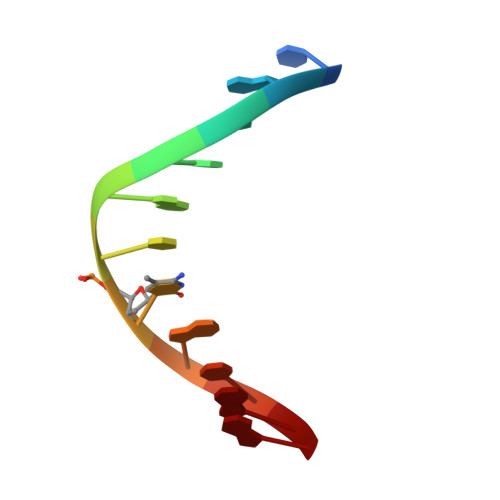
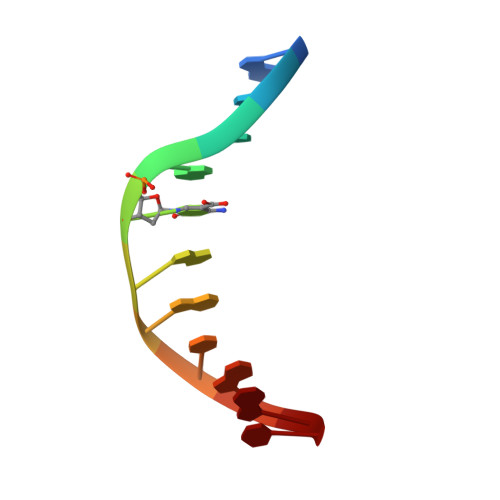
DNA recognition of 5-carboxylcytosine by a zfp57 mutant at an atomic resolution of 0.97 angstrom.
Liu, Y., Olanrewaju, Y.O., Zhang, X., Cheng, X.(2013) Biochemistry 52: 9310-9317
- PubMed: 24236546
- DOI: https://doi.org/10.1021/bi401360n
- Primary Citation of Related Structures:
4M9V - PubMed Abstract:
The Zfp57 gene encodes a KRAB (Krüppel-associated box) domain-containing C2H2 zinc finger transcription factor that is expressed in early development. Zfp57 protein recognizes methylated CpG dinucleotide within GCGGCA elements at multiple imprinting control regions. In the previously determined structure of the mouse Zfp57 DNA-binding domain in complex with DNA containing 5-methylcytosine (5mC), the side chains of Arg178 and Glu182 contact the methyl group via hydrophobic and van der Waals interactions. We examined the role of Glu182 in recognition of 5mC by mutagenesis. The majority of mutants examined lose selectivity of methylated (5mC) over unmodified (C) and oxidative derivatives, 5-hydroxymethylcytosine, 5-formylcytosine, and 5-carboxylcytosine (5caC), suggesting that the side chain of Glu182 (the size and the charge) is dispensable for methyl group recognition but negatively impacts the binding of unmodified cytosine as well as oxidized derivatives of 5mC to achieve 5mC selectivity. Substitution of Glu182 with its corresponding amide (E182Q) had no effect on methylated DNA binding but gained significant binding affinity for 5caC DNA, resulting in a binding affinity for 5caC DNA comparable to that of the wild-type protein for 5mC. We show structurally that the uncharged amide group of E182Q interacts favorably with the carboxylate group of 5caC. Furthermore, introducing a positively charged arginine at position 182 resulted in a mutant (E182R) having higher selectivity for the negatively charged 5caC.
- Department of Biochemistry, Emory University School of Medicine , 1510 Clifton Road, Atlanta, Georgia 30322, United States.
Organizational Affiliation: