Structural basis for activation and non-canonical catalysis of the Rap GTPase activating protein domain of plexin.
Wang, Y., Pascoe, H.G., Brautigam, C.A., He, H., Zhang, X.(2013) Elife 2: e01279-e01279
- PubMed: 24137545
- DOI: https://doi.org/10.7554/eLife.01279
- Primary Citation of Related Structures:
4M8M, 4M8N - PubMed Abstract:
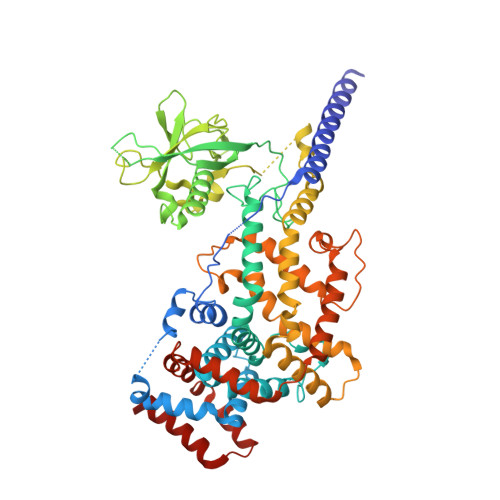
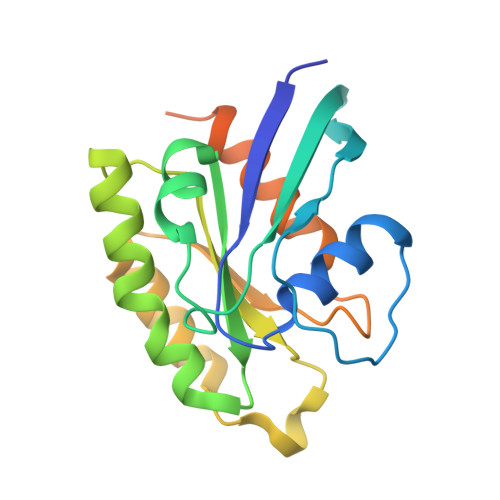
Plexins are cell surface receptors that bind semaphorins and transduce signals for regulating neuronal axon guidance and other processes. Plexin signaling depends on their cytoplasmic GTPase activating protein (GAP) domain, which specifically inactivates the Ras homolog Rap through an ill-defined non-canonical catalytic mechanism. The plexin GAP is activated by semaphorin-induced dimerization, the structural basis for which remained unknown. Here we present the crystal structures of the active dimer of zebrafish PlexinC1 cytoplasmic region in the apo state and in complex with Rap. The structures show that the dimerization induces a large-scale conformational change in plexin, which opens the GAP active site to allow Rap binding. Plexin stabilizes the switch II region of Rap in an unprecedented conformation, bringing Gln63 in Rap into the active site for catalyzing GTP hydrolysis. The structures also explain the unique Rap-specificity of plexins. Mutational analyses support that these mechanisms underlie plexin activation and signaling. DOI:http://dx.doi.org/10.7554/eLife.01279.001.
- Department of Pharmacology , University of Texas Southwestern Medical Center , Dallas , United States.
Organizational Affiliation: