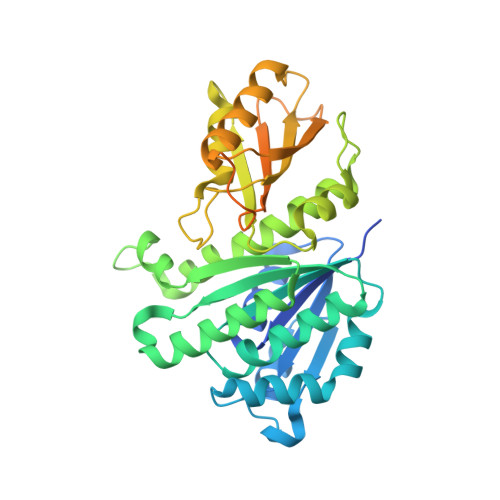
1.43 Angstrom resolution crystal structure of cell division protein FtsZ (ftsZ) from Staphylococcus epidermidis RP62A in complex with GDP
Halavaty, A.S., Minasov, G., Winsor, J., Dubrovska, I., Filippova, E.V., Olsen, D.B., Therien, A., Shuvalova, L., Young, K., Anderson, W.F.To be published.