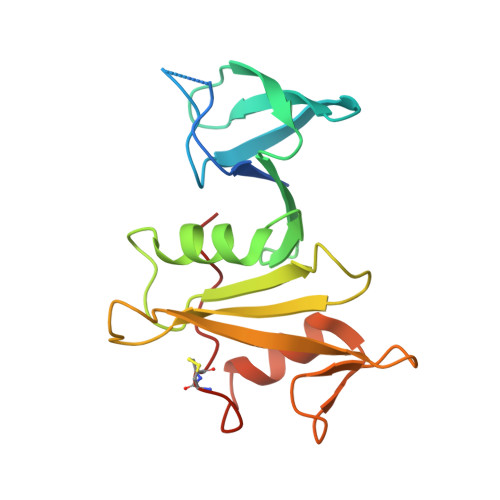
Crystal structure of Src-like adaptor protein 2 reveals close association of SH3 and SH2 domains through beta-sheet formation.
Wybenga-Groot, L.E., McGlade, C.J.(2013) Cell Signal 25: 2702-2708
- PubMed: 24018043
- DOI: https://doi.org/10.1016/j.cellsig.2013.08.040
- Primary Citation of Related Structures:
4M4Z - PubMed Abstract:
The Src-like adaptor proteins (SLAP/SLAP2) are key components of Cbl-dependent downregulation of antigen receptor, cytokine receptor, and receptor tyrosine kinase signaling in hematopoietic cells. SLAP and SLAP2 consist of adjacent SH3 and SH2 domains that are most similar in sequence to Src family kinases (SFKs). Notably, the SH3-SH2 connector sequence is significantly shorter in SLAP/SLAP2 than in SFKs. To understand the structural implication of a short SH3-SH2 connector sequence, we solved the crystal structure of a protein encompassing the SH3 domain, SH3-SH2 connector, and SH2 domain of SLAP2 (SLAP2-32). While both domains adopt typical folds, the short SH3-SH2 connector places them in close association. Strand βe of the SH3 domain interacts with strand βA of the SH2 domain, resulting in the formation of a continuous β sheet that spans the length of the protein. Disruption of the SH3/SH2 interface through mutagenesis decreases SLAP-32 stability in vitro, consistent with inter-domain binding being an important component of SLAP2 structure and function. The canonical peptide binding pockets of the SH3 and SH2 domains are fully accessible, in contrast to other protein structures that display direct interaction between SH3 and SH2 domains, in which either peptide binding surface is obstructed by the interaction. Our results reveal potential sites of novel interaction for SH3 and SH2 domains, and illustrate the adaptability of SH2 and SH3 domains in mediating interactions. As well, our results suggest that the SH3 and SH2 domains of SLAP2 function interdependently, with implications on their mode of substrate binding.
- The Arthur and Sonia Labatt Brain Tumour Research Centre and Program in Cell Biology, The Hospital for Sick Children, 555 University Avenue, Toronto, ON M5G 1X8, Canada. Electronic address: leanne.wybenga.groot@utoronto.ca.
Organizational Affiliation: