Molecular Dynamic Studies of Reaction Centers Mutants from Rhodobacter sphaeroides and his mutant form L(M196)H+H(M202)L
Kljashtorny, V.G., Fufina, T.Y., Vasilieva, L.G., Gabdulkhakov, A.G.(2014) Crystallogr Rep 59: 536-541
Experimental Data Snapshot
Starting Model: experimental
View more details
(2014) Crystallogr Rep 59: 536-541
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Reaction center protein H chain | A [auth H] | 260 | Cereibacter sphaeroides | Mutation(s): 0 Gene Names: puhA Membrane Entity: Yes | 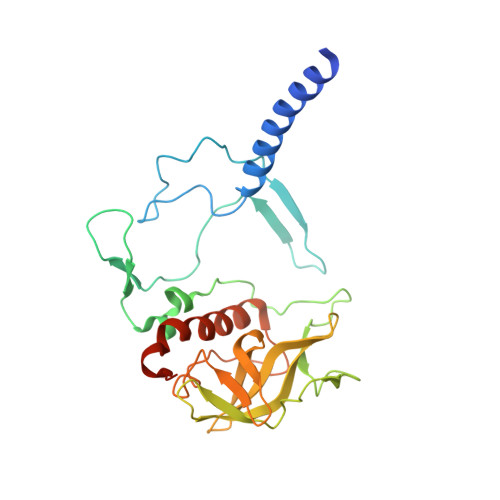 |
UniProt | |||||
Find proteins for P0C0Y7 (Cereibacter sphaeroides) Explore P0C0Y7 Go to UniProtKB: P0C0Y7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0C0Y7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Reaction center protein L chain | B [auth L] | 282 | Cereibacter sphaeroides | Mutation(s): 0 Gene Names: pufL Membrane Entity: Yes | 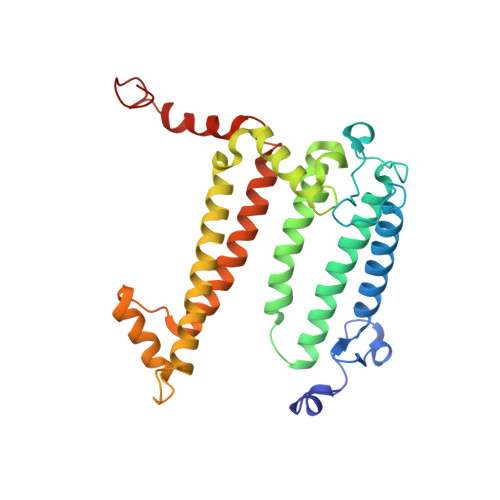 |
UniProt | |||||
Find proteins for P0C0Y8 (Cereibacter sphaeroides) Explore P0C0Y8 Go to UniProtKB: P0C0Y8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0C0Y8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Reaction center protein M chain | C [auth M] | 303 | Cereibacter sphaeroides | Mutation(s): 2 Gene Names: pufM Membrane Entity: Yes | 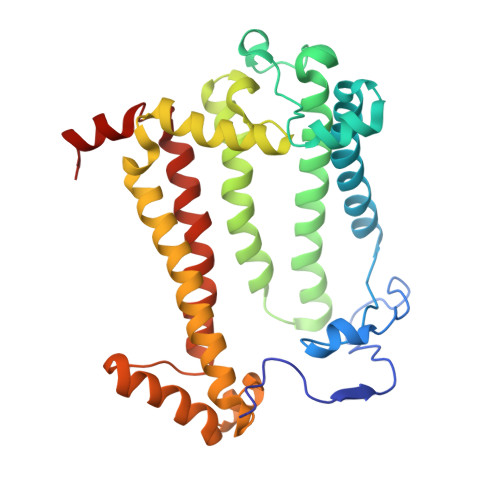 |
UniProt | |||||
Find proteins for P0C0Y9 (Cereibacter sphaeroides) Explore P0C0Y9 Go to UniProtKB: P0C0Y9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0C0Y9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 13 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CDL Query on CDL | CA [auth M] | CARDIOLIPIN C81 H156 O17 P2 XVTUQDWPJJBEHJ-KZCWQMDCSA-L |  | ||
| BCL Query on BCL | EA [auth M], Q [auth L], R [auth L] | BACTERIOCHLOROPHYLL A C55 H74 Mg N4 O6 DSJXIQQMORJERS-AGGZHOMASA-M |  | ||
| BPH Query on BPH | DA [auth M], FA [auth M], S [auth L] | BACTERIOPHEOPHYTIN A C55 H76 N4 O6 KWOZSBGNAHVCKG-SZQBJALDSA-N |  | ||
| U10 Query on U10 | HA [auth M], T [auth L] | UBIQUINONE-10 C59 H90 O4 ACTIUHUUMQJHFO-UPTCCGCDSA-N |  | ||
| SPN Query on SPN | IA [auth M] | SPEROIDENONE C41 H70 O2 GWQAMGYOEYXWJF-YCDPMLDASA-N |  | ||
| LDA Query on LDA | D [auth H] JA [auth M] KA [auth M] LA [auth M] MA [auth M] | LAURYL DIMETHYLAMINE-N-OXIDE C14 H31 N O SYELZBGXAIXKHU-UHFFFAOYSA-N |  | ||
| TRS Query on TRS | BA [auth L], O [auth H] | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL C4 H12 N O3 LENZDBCJOHFCAS-UHFFFAOYSA-O |  | ||
| PO4 Query on PO4 | I [auth H], RA [auth M], SA [auth M] | PHOSPHATE ION O4 P NBIIXXVUZAFLBC-UHFFFAOYSA-K |  | ||
| DIO Query on DIO | J [auth H], TA [auth M], Y [auth L], Z [auth L] | 1,4-DIETHYLENE DIOXIDE C4 H8 O2 RYHBNJHYFVUHQT-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | AA [auth L] K [auth H] L [auth H] M [auth H] N [auth H] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| FE Query on FE | GA [auth M] | FE (III) ION Fe VTLYFUHAOXGGBS-UHFFFAOYSA-N |  | ||
| K Query on K | P [auth H] | POTASSIUM ION K NPYPAHLBTDXSSS-UHFFFAOYSA-N |  | ||
| UNL Query on UNL | E [auth H] F [auth H] G [auth H] H OA [auth M] | Unknown ligand NPYPAHLBTDXSSS-UHFFFAOYSA-N | |||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 140.04 | α = 90 |
| b = 140.04 | β = 90 |
| c = 184.62 | γ = 120 |
| Software Name | Purpose |
|---|---|
| XDS | data scaling |
| PHASER | phasing |
| PHENIX | refinement |
| XDS | data reduction |