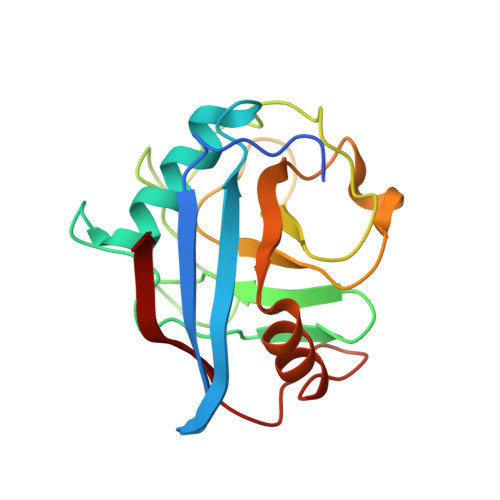
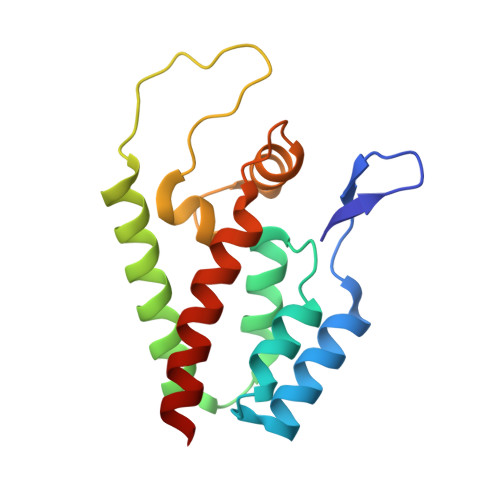
HIV-1 capsid undergoes coupled binding and isomerization by the nuclear pore protein NUP358.
Bichel, K., Price, A.J., Schaller, T., Towers, G.J., Freund, S.M., James, L.C.(2013) Retrovirology 10: 81-81
- PubMed: 23902822
- DOI: https://doi.org/10.1186/1742-4690-10-81
- Primary Citation of Related Structures:
4LQW - PubMed Abstract:
Lentiviruses such as HIV-1 can be distinguished from other retroviruses by the cyclophilin A-binding loop in their capsid and their ability to infect non-dividing cells. Infection of non-dividing cells requires transport through the nuclear pore but how this is mediated is unknown. Here we present the crystal structure of the N-terminal capsid domain of HIV-1 in complex with the cyclophilin domain of nuclear pore protein NUP358. The structure reveals that HIV-1 is positioned to allow single-bond resonance stabilisation of exposed capsid residue P90. NMR exchange experiments demonstrate that NUP358 is an active isomerase, which efficiently catalyzes cis-trans isomerization of the HIV-1 capsid. In contrast, the distantly related feline lentivirus FIV can bind NUP358 but is neither isomerized by it nor requires it for infection. Isomerization by NUP358 may be preserved by HIV-1 to target the nuclear pore and synchronize nuclear entry with capsid uncoating.
- Protein and Nucleic Acid Chemistry Division, Medical Research Council Laboratory of Molecular Biology, Hills Road, Cambridge CB2 0QH, UK.
Organizational Affiliation: