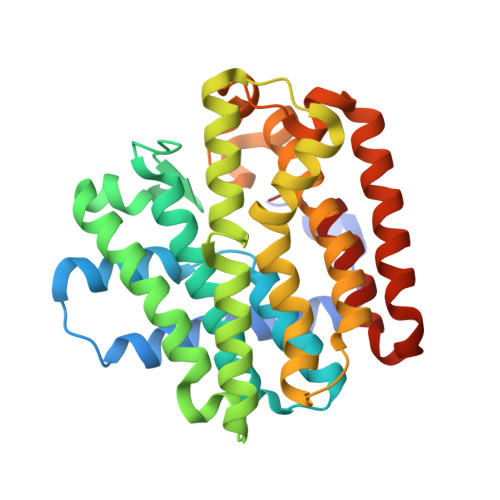
Crystal structure of a farnesyl diphosphate synthase from Roseobacter denitrificans OCh 114, target EFI-509393, with two IPP and calcium bound in active site
Kim, J., Toro, R., Bhosle, R., Al Obaidi, N.F., Morisco, L.L., Wasserman, S.R., Sojitra, S., Washington, E., Scott Glenn, A., Chowdhury, S., Evans, B., Hammonds, J., Hillerich, B., Love, J., Seidel, R.D., Imker, H.J., Stead, M., Gerlt, J.A., Almo, S.C.To be published.