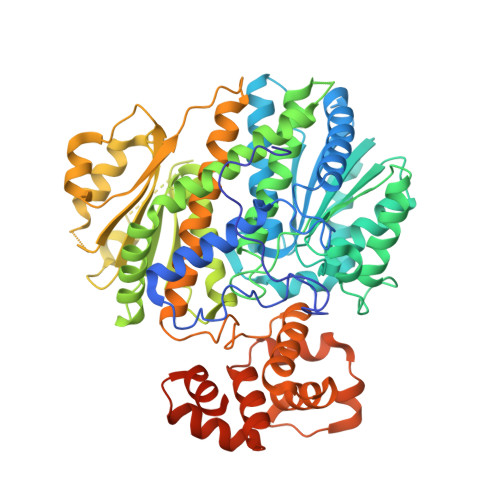
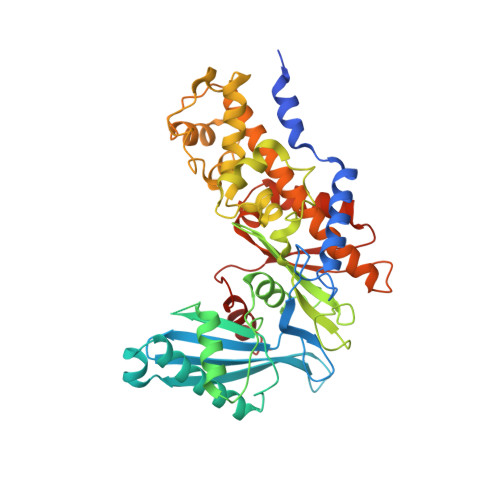
Structural basis for regulation of human glucokinase by glucokinase regulatory protein.
Beck, T., Miller, B.G.(2013) Biochemistry 52: 6232-6239
- PubMed: 23957911
- DOI: https://doi.org/10.1021/bi400838t
- Primary Citation of Related Structures:
4LC9 - PubMed Abstract:
Glucokinase (GCK) is responsible for maintaining glucose homeostasis in the human body. Dysfunction or misregulation of GCK causes hyperinsulinemia, hypertriglyceridemia, and type 2 diabetes. In the liver, GCK is regulated by interaction with the glucokinase regulatory protein (GKRP), a 68 kDa polypeptide that functions as a competitive inhibitor of glucose binding to GCK. Formation of the mammalian GCK-GKRP complex is stimulated by fructose 6-phosphate and antagonized by fructose 1-phosphate. Here we report the crystal structure of the mammalian GCK-GKRP complex in the presence of fructose 6-phosphate at a resolution of 3.50 Å. The interaction interface, which totals 2060 Å(2) of buried surface area, is characterized by a small number of polar contacts and substantial hydrophobic interactions. The structure of the complex reveals the molecular basis of disease states associated with impaired regulation of GCK by GKRP. It also offers insight into the modulation of complex stability by sugar phosphates. The atomic description of the mammalian GCK-GKRP complex provides a framework for the development of novel diabetes therapeutic agents that disrupt this critical macromolecular regulatory unit.
- Laboratory of Organic Chemistry, ETH Zürich , Zürich CH-8093, Switzerland.
Organizational Affiliation: