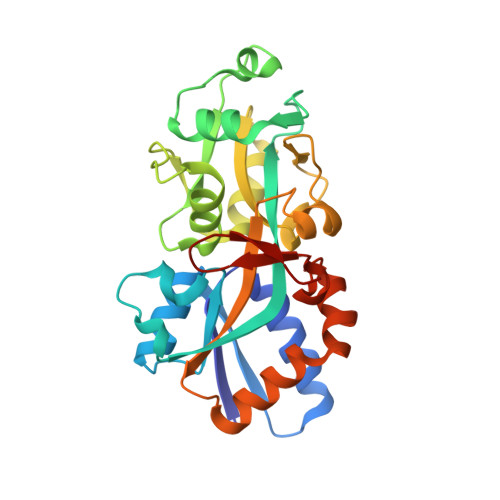
Crystal structure of phosphate ABC transporter, periplasmic phosphate-binding protein PstS 1 (PBP1) from Streptococcus pneumoniae Canada MDR_19A in complex with phosphate
Stogios, P.J., Wawrzak, Z., Kudritska, M., Yim, V., Savchenko, A., Anderson, W.F.To be published.