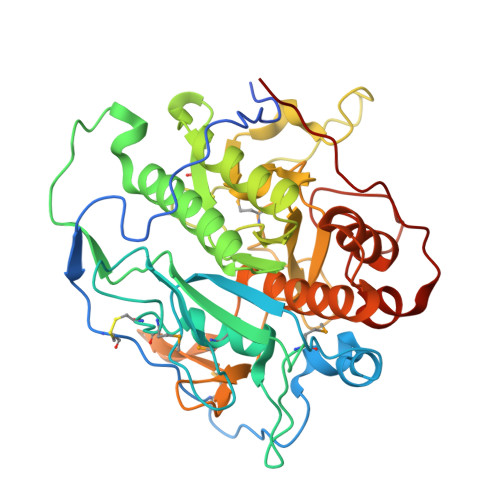
Structure of the Mycosin-1 Protease from the Mycobacterial ESX-1 Protein Type VII Secretion System.
Solomonson, M., Huesgen, P.F., Wasney, G.A., Watanabe, N., Gruninger, R.J., Prehna, G., Overall, C.M., Strynadka, N.C.(2013) J Biological Chem 288: 17782-17790
- PubMed: 23620593
- DOI: https://doi.org/10.1074/jbc.M113.462036
- Primary Citation of Related Structures:
4J94, 4KPG - PubMed Abstract:
Mycobacteria use specialized type VII (ESX) secretion systems to export proteins across their complex cell walls. Mycobacterium tuberculosis encodes five nonredundant ESX secretion systems, with ESX-1 being particularly important to disease progression. All ESX loci encode extracellular membrane-bound proteases called mycosins (MycP) that are essential to secretion and have been shown to be involved in processing of type VII-exported proteins. Here, we report the first x-ray crystallographic structure of MycP1(24-407) to 1.86 Å, defining a subtilisin-like fold with a unique N-terminal extension previously proposed to function as a propeptide for regulation of enzyme activity. The structure reveals that this N-terminal extension shows no structural similarity to previously characterized protease propeptides and instead wraps intimately around the catalytic domain where, tethered by a disulfide bond, it forms additional interactions with a unique extended loop that protrudes from the catalytic core. We also show MycP1 cleaves the ESX-1 secreted protein EspB from both M. tuberculosis and Mycobacterium smegmatis at a homologous cut site in vitro.
- Department of Biochemistry and Molecular Biology and Centre for Blood Research, University of British Columbia, Vancouver, British Columbia V6T 1Z3, Canada.
Organizational Affiliation: