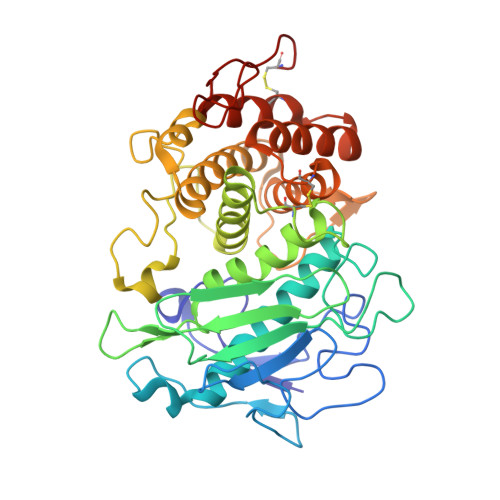
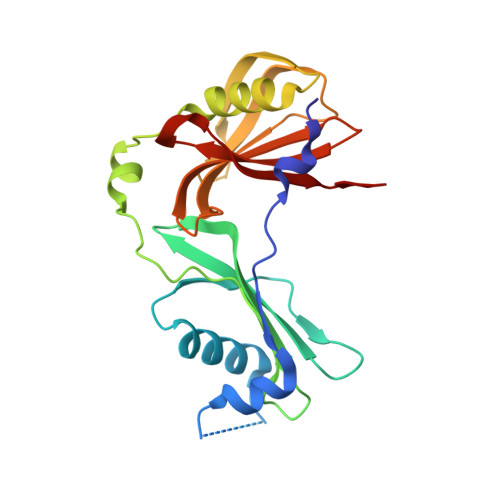
A functional and structural study of the major metalloprotease secreted by the pathogenic fungus Aspergillus fumigatus.
Fernandez, D., Russi, S., Vendrell, J., Monod, M., Pallares, I.(2013) Acta Crystallogr D Biol Crystallogr 69: 1946-1957
- PubMed: 24100314
- DOI: https://doi.org/10.1107/S0907444913017642
- Primary Citation of Related Structures:
4K90 - PubMed Abstract:
Fungalysins are secreted fungal peptidases with the ability to degrade the extracellular matrix proteins elastin and collagen and are thought to act as virulence factors in diseases caused by fungi. Fungalysins constitute a unique family among zinc-dependent peptidases that bears low sequence similarity to known bacterial peptidases of the thermolysin family. The crystal structure of the archetype of the fungalysin family, Aspergillus fumigatus metalloprotease (AfuMep), has been obtained for the first time. The 1.8 Å resolution structure of AfuMep corresponds to that of an autoproteolyzed proenzyme with separate polypeptide chains corresponding to the N-terminal prodomain in a binary complex with the C-terminal zinc-bound catalytic domain. The prodomain consists of a tandem of cystatin-like folds whose C-terminal end is buried into the active-site cleft of the catalytic domain. The catalytic domain harbouring the key catalytic zinc ion and its ligands, two histidines and one glutamic acid, undergoes a conspicuous rearrangement of its N-terminal end during maturation. One key positively charged amino-acid residue and the C-terminal disulfide bridge appear to contribute to its structural-functional properties. Thus, structural, biophysical and biochemical analysis were combined to provide a deeper comprehension of the underlying properties of A. fumigatus fungalysin, serving as a framework for the as yet poorly known metallopeptidases from pathogenic fungi.
- Departament de Bioquímica i Biologia Molecular, Facultat de Biociències, Universitat Autònoma de Barcelona, 08193 Bellaterra, Spain.
Organizational Affiliation: