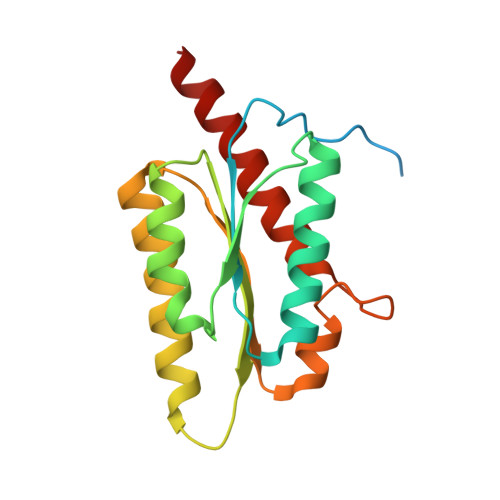
Crystal structure of a Probable Riboflavin Synthase, Beta chain RIBH (6,7-dimethyl-8-ribityllumazine synthase, DMRL synthase, Lumazine synthase) from Mycobacterium leprae
Seattle Structural Genomics Center for Infectious Disease (SSGCID), Abendroth, J., Lorimer, D., Edwards, T.E.To be published.