Fully Convergent Chemical Synthesis of Ester Insulin: Determination of the High Resolution X-ray Structure by Racemic Protein Crystallography.
Avital-Shmilovici, M., Mandal, K., Gates, Z.P., Phillips, N.B., Weiss, M.A., Kent, S.B.(2013) J Am Chem Soc 135: 3173-3185
- PubMed: 23343390
- DOI: https://doi.org/10.1021/ja311408y
- Primary Citation of Related Structures:
4IUZ - PubMed Abstract:
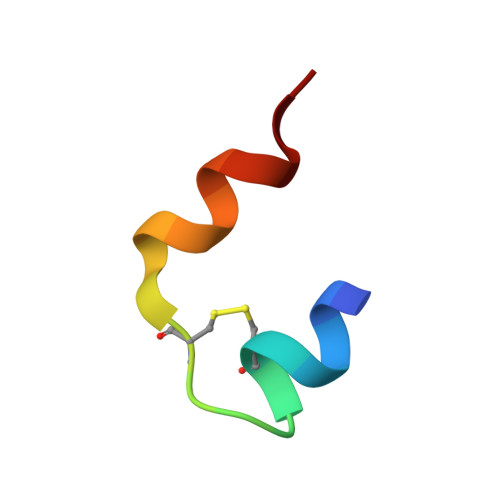
Efficient total synthesis of insulin is important to enable the application of medicinal chemistry to the optimization of the properties of this important protein molecule. Recently we described "ester insulin"--a novel form of insulin in which the function of the 35 residue C-peptide of proinsulin is replaced by a single covalent bond--as a key intermediate for the efficient total synthesis of insulin. Here we describe a fully convergent synthetic route to the ester insulin molecule from three unprotected peptide segments of approximately equal size. The synthetic ester insulin polypeptide chain folded much more rapidly than proinsulin, and at physiological pH. Both the D-protein and L-protein enantiomers of monomeric DKP ester insulin (i.e., [Asp(B10), Lys(B28), Pro(B29)]ester insulin) were prepared by total chemical synthesis. The atomic structure of the synthetic ester insulin molecule was determined by racemic protein X-ray crystallography to a resolution of 1.6 Å. Diffraction quality crystals were readily obtained from the racemic mixture of {D-DKP ester insulin + L-DKP ester insulin}, whereas crystals were not obtained from the L-ester insulin alone even after extensive trials. Both the D-protein and L-protein enantiomers of monomeric DKP ester insulin were assayed for receptor binding and in diabetic rats, before and after conversion by saponification to the corresponding DKP insulin enantiomers. L-DKP ester insulin bound weakly to the insulin receptor, while synthetic L-DKP insulin derived from the L-DKP ester insulin intermediate was fully active in binding to the insulin receptor. The D- and L-DKP ester insulins and D-DKP insulin were inactive in lowering blood glucose in diabetic rats, while synthetic L-DKP insulin was fully active in this biological assay. The structural basis of the lack of biological activity of ester insulin is discussed.
- Department of Chemistry, The University of Chicago, Chicago, Illinois 60637, USA.
Organizational Affiliation: