Structural insight into plant programmed cell death mediated by BAG proteins in Arabidopsis thaliana.
Fang, S., Li, L., Cui, B., Men, S., Shen, Y., Yang, X.(2013) Acta Crystallogr D Biol Crystallogr 69: 934-945
- PubMed: 23695238
- DOI: https://doi.org/10.1107/S0907444913003624
- Primary Citation of Related Structures:
4HWC, 4HWD, 4HWF, 4HWH, 4HWI - PubMed Abstract:
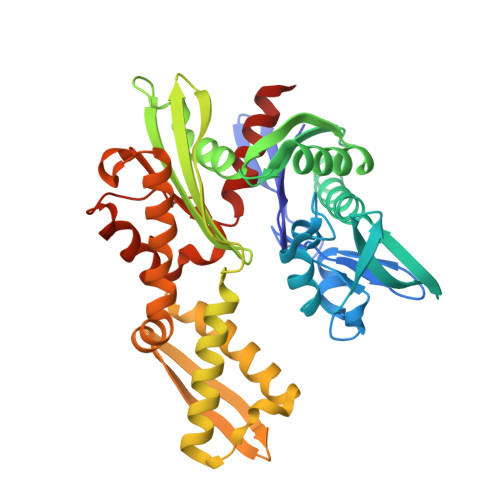
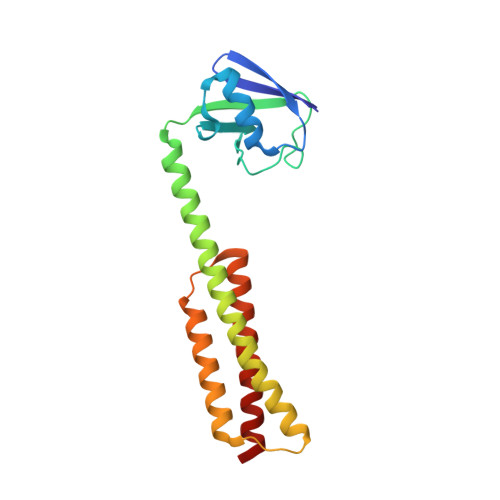
The recently identified plant Bcl-2-associated athanogene (BAG) family plays an extensive role in plant programmed cell death (PCD) processes ranging from growth and development to stress responses and even cell death. In the Arabidopsis thaliana BAG (AtBAG) protein family, four members (AtBAG1-4) have a domain organization similar to that of mammalian BAG proteins. Here, crystal structures of the BAG domains (BDs) of AtBAG1-4 have been determined; they have high homology and adopt a structure comprising three short parallel α-helices, similar to some mammalian BAG proteins. The crystal structure of a complex of the AtBAG1 ubiquitin-like domain and BAG domain (UBD) with the Hsc70 nucleotide-binding domain (NBD) was also determined. The binding of the AtBAG1 BD to the Hsc70 NBD induces conformational change of the Hsc70 NBD to the open state and reduces the affinity of the NBD for ADP. In vivo studies showed that bag2-1 mutant plants are larger than wild-type plants when growing under normal conditions, indicating that the AtBAG proteins might regulate plant PCD and confer tolerance to stresses in plants. These structural and functional analyses indicate that the AtBAG proteins function as nucleotide-exchange factors for Hsp70/Hsc70 in A. thaliana and that the mechanism of regulation of chaperone-mediated protein folding is conserved in plants.
- State Key Laboratory of Medicinal Chemical Biology, Nankai University, Tianjin 300071, People's Republic of China.
Organizational Affiliation: