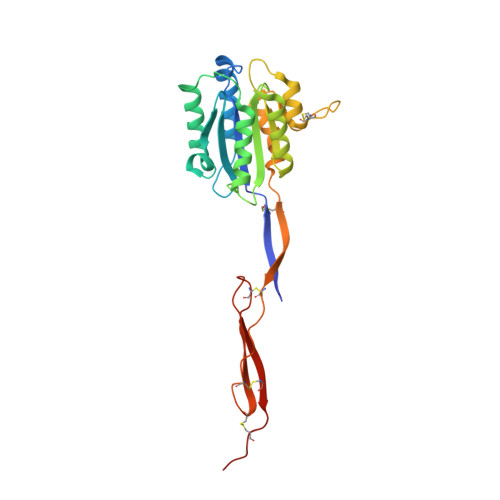
Shape change in the receptor for gliding motility in Plasmodium sporozoites.
Song, G., Koksal, A.C., Lu, C., Springer, T.A.(2012) Proc Natl Acad Sci U S A 109: 21420-21425
- PubMed: 23236185
- DOI: https://doi.org/10.1073/pnas.1218581109
- Primary Citation of Related Structures:
4HQF, 4HQK, 4HQL, 4HQN, 4HQO - PubMed Abstract:
Sporozoite gliding motility and invasion of mosquito and vertebrate host cells in malaria is mediated by thrombospondin repeat anonymous protein (TRAP). Tandem von Willebrand factor A (VWA) and thrombospondin type I repeat (TSR) domains in TRAP connect through proline-rich stalk, transmembrane, and cytoplasmic domains to the parasite actin-dependent motility apparatus. We crystallized fragments containing the VWA and TSR domains from Plasmodium vivax and Plasmodium falciparum in different crystal lattices. TRAP VWA domains adopt closed and open conformations, and bind a Mg(2+) ion at a metal ion-dependent adhesion site implicated in ligand binding. Metal ion coordination in the open state is identical to that seen in the open high-affinity state of integrin I domains. The closed VWA conformation associates with a disordered TSR domain. In contrast, the open VWA conformation crystallizes with an extensible β ribbon and ordered TSR domain. The extensible β ribbon is composed of disulfide-bonded segments N- and C-terminal to the VWA domain that are largely drawn out of the closed VWA domain in a 15 Å movement to the open conformation. The extensible β ribbon and TSR domain overlap at a conserved interface. The VWA, extensible β ribbon, and TSR domains adopt a highly elongated overall orientation that would be stabilized by tensile force exerted across a ligand-receptor complex by the actin motility apparatus of the sporozoite. Our results provide insights into regulation of "stick-and-slip" parasite motility and for development of sporozoite subunit vaccines.
- Immune Disease Institute, Program in Cellular and Molecular Medicine, Children's Hospital Boston, MA 02115, USA.
Organizational Affiliation: