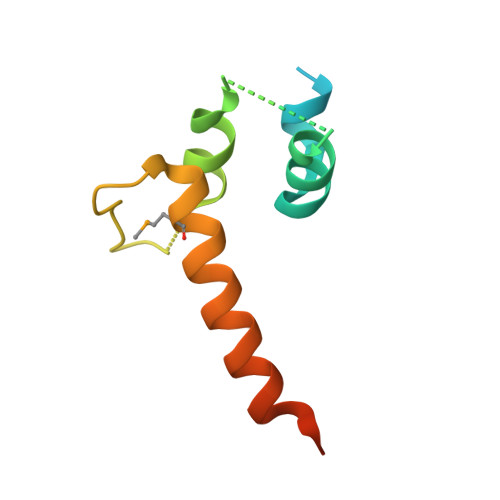
Crystal structure of a transcription elongation factor B polypeptide 3 from Homo sapiens, Northeast Structural Genomics consortium target id HR4748B. (CASP Target)
Seetharaman, J., Su, M., Ciccosanti, C., Sahdev, S., Acton, T.B., Xiao, R., K Everett, J., T Montelione, G., Hunt, J.F., Tong, L.To be published.