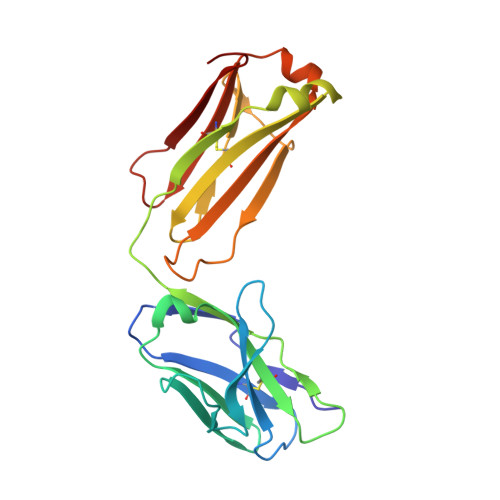
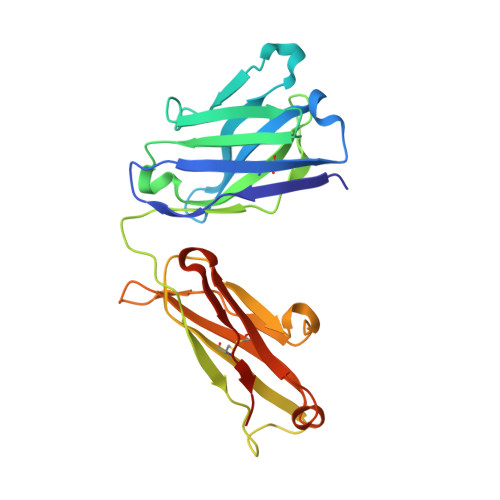
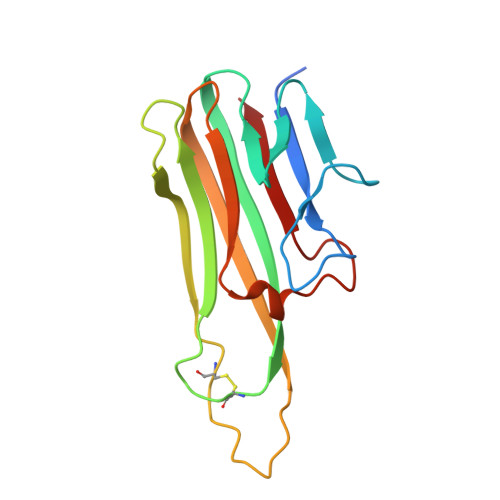
Structural basis for treating tumor necrosis factor alpha (TNFalpha)-associated diseases with the therapeutic antibody infliximab
Liang, S.Y., Dai, J.X., Hou, S., Su, L., Zhang, D., Guo, H., Hu, S., Wang, H., Rao, Z., Guo, Y.J., Lou, Z.Y.(2013) J Biological Chem 288: 13799-13807
- PubMed: 23504311
- DOI: https://doi.org/10.1074/jbc.M112.433961
- Primary Citation of Related Structures:
4G3Y - PubMed Abstract:
Although infliximab has high efficacy in treating TNFα-associated diseases, the epitope on TNFα remains unclear. The crystal structure of the TNFα in complex with the infliximab Fab is reported at a resolution of 2.6 Å. TNFα E-F loop plays a crucial role in the interaction. The structure may lead to understanding the mechanism of mAb anti-TNFα Monoclonal antibody (mAb) drugs have been widely used for treating tumor necrosis factor α (TNFα)-related diseases for over 10 years. Although their action has been hypothesized to depend in part on their ability to bind precursor cell surface TNFα, the precise mechanism and the epitope bound on TNFα remain unclear. In the present work, we report the crystal structure of the infliximab Fab fragment in complex with TNFα at a resolution of 2.6 Å. The key features of the TNFα E-F loop region in this complex distinguish the interaction between infliximab and TNFα from other TNF-receptor structures, revealing the mechanism of TNFα inhibition by overlapping with the TNFα-receptor interface and indicating the crucial role of the E-F loop in the action of this therapeutic antibody. This structure also indicates the formation of an aggregated network for the activation of complement-dependent cytolysis and antibody-dependent cell-mediated cytotoxicity, which result in development of granulomatous infections through TNFα blockage. These results provide the first experimental model for the interaction of TNFα with therapeutic antibodies and offer useful information for antibody optimization by understanding the precise molecular mechanism of TNFα inhibition.
- Laboratory of Structural Biology and Ministry of Education Laboratory of Protein Science, School of Medicine, Tsinghua University, Beijing 100084, China.
Organizational Affiliation: