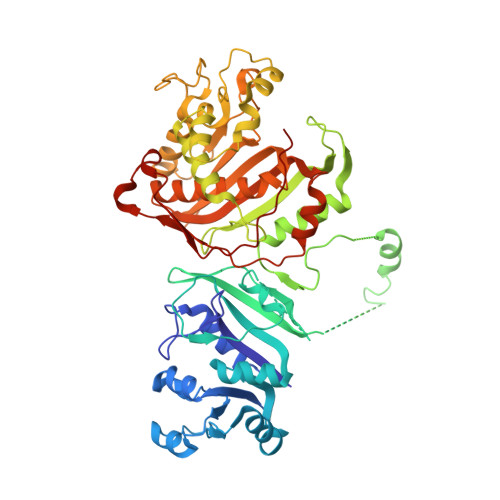
First Three-Dimensional Structure of Toxoplasma gondii Thymidylate Synthase-Dihydrofolate Reductase: Insights for Catalysis, Interdomain Interactions, and Substrate Channeling.
Sharma, H., Landau, M.J., Vargo, M.A., Spasov, K.A., Anderson, K.S.(2013) Biochemistry 52: 7305-7317
- PubMed: 24053355
- DOI: https://doi.org/10.1021/bi400576t
- Primary Citation of Related Structures:
4ECK, 4EIL - PubMed Abstract:
Most species, such as humans, have monofunctional forms of thymidylate synthase (TS) and dihydrofolate reductase (DHFR) that are key folate metabolism enzymes making critical folate components required for DNA synthesis. In contrast, several parasitic protozoa, including Toxoplasma gondii , contain a unique bifunctional thymidylate synthase-dihydrofolate reductase (TS-DHFR) having the catalytic activities contained on a single polypeptide chain. The prevalence of T. gondii infections across the world, especially for those immunocompromised, underscores the need to understand TS-DHFR enzyme function and to find new avenues to exploit for the design of novel antiparasitic drugs. As a first step, we have solved the first three-dimensional structures of T. gondii TS-DHFR at 3.7 Å and of a loop truncated TS-DHFR, removing several flexible surface loops in the DHFR domain, improving resolution to 2.2 Å. Distinct structural features of the TS-DHFR homodimer include a junctional region containing a kinked crossover helix between the DHFR domains of the two adjacent monomers, a long linker connecting the TS and DHFR domains, and a DHFR domain that is positively charged. The roles of these unique structural features were probed by site-directed mutagenesis coupled with presteady state and steady state kinetics. Mutational analysis of the crossover helix region combined with kinetic characterization established the importance of this region not only in DHFR catalysis but also in modulating the distal TS activity, suggesting a role for TS-DHFR interdomain interactions. Additional kinetic studies revealed that substrate channeling occurs in which dihydrofolate is directly transferred from the TS to DHFR active site without entering bulk solution. The crystal structure suggests that the positively charged DHFR domain governs this electrostatically mediated movement of dihydrofolate, preventing release from the enzyme. Taken together, these structural and kinetic studies reveal unique, functional regions on the T. gondii TS-DHFR enzyme that may be targeted for inhibition, thus paving the way for designing species specific inhibitors.
- The Department of Pharmacology, Yale University School of Medicine, New Haven, CT 06510.
Organizational Affiliation: