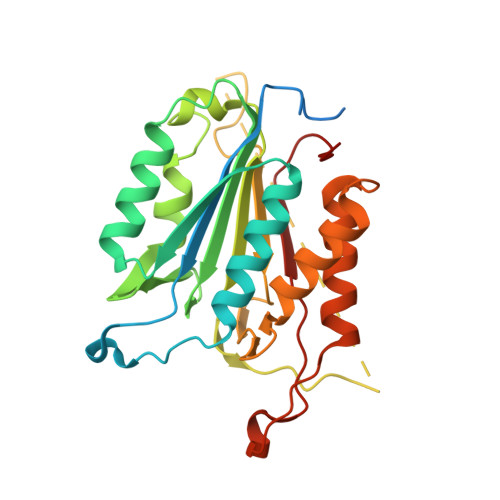
Allosteric modulation of caspase 3 through mutagenesis.
Walters, J., Schipper, J.L., Swartz, P., Mattos, C., Clark, A.C.(2012) Biosci Rep 32: 401-411
- PubMed: 22607239
- DOI: https://doi.org/10.1042/BSR20120037
- Primary Citation of Related Structures:
4EHA, 4EHD, 4EHF, 4EHH, 4EHK, 4EHL, 4EHN - PubMed Abstract:
A mutation in the allosteric site of the caspase 3 dimer interface of Val266 to histidine abolishes activity of the enzyme, and models predict that the mutation mimics the action of small molecule allosteric inhibitors by preventing formation of the active site. Mutations were coupled to His266 at two sites in the interface, E124A and Y197C. We present results from X-ray crystallography, enzymatic activity and molecular dynamics simulations for seven proteins, consisting of single, double and triple mutants. The results demonstrate that considering allosteric inhibition of caspase 3 as a shift between discrete 'off-state' or 'on-state' conformations is insufficient. Although His266 is accommodated in the interface, the structural defects are propagated to the active site through a helix on the protein surface. A more comprehensive view of allosteric regulation of caspase 3 requires the representation of an ensemble of inactive states and shows that subtle structural changes lead to the population of the inactive ensemble.
- Department of Molecular and Structural Biochemistry, North Carolina State University, Raleigh, NC 27695, USA.
Organizational Affiliation: