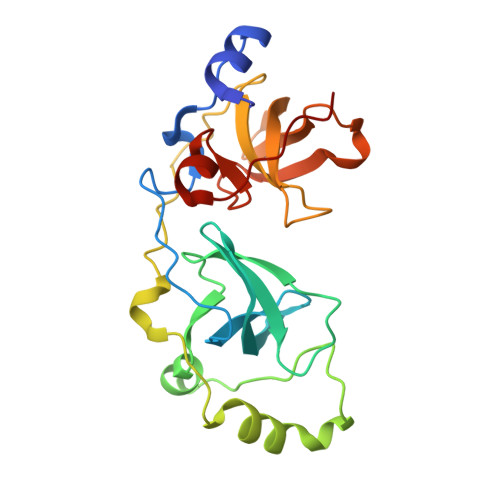
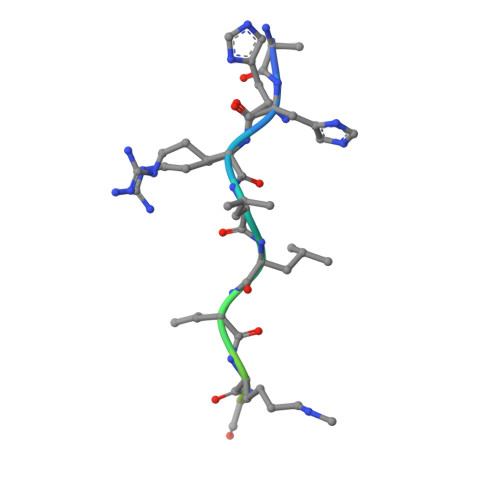
Histone recognition by human malignant brain tumor domains.
Nady, N., Krichevsky, L., Zhong, N., Duan, S., Tempel, W., Amaya, M.F., Ravichandran, M., Arrowsmith, C.H.(2012) J Mol Biology 423: 702-718
- PubMed: 22954662
- DOI: https://doi.org/10.1016/j.jmb.2012.08.022
- Primary Citation of Related Structures:
4EDU - PubMed Abstract:
Histone methylation has emerged as an important covalent modification involved in a variety of biological processes, especially regulation of transcription and chromatin dynamics. Lysine methylation is found in three distinct states (monomethylation, dimethylation and trimethylation), which are recognized by specific protein domains. The malignant brain tumor (MBT) domain is one such module found in several chromatin regulatory complexes including Polycomb repressive complex 1. Here, we present a comprehensive characterization of the human MBT family with emphasis on histone binding specificity. SPOT-blot peptide arrays were used to screen for the methyllysine-containing histone peptides that bind to MBT domains found in nine human proteins. Selected interactions were quantified using fluorescence polarization assays. We show that all MBT proteins recognize only monomethyllysine and/or dimethyllysine marks and provide evidence that some MBT domains recognize a defined consensus sequence while others bind in a promiscuous, non-sequence-specific manner. Furthermore, using structure-based mutants, we identify a triad of residues in the methyllysine binding pocket that imparts discrimination between monomethyllysine and dimethyllysine. This study represents a comprehensive analysis of MBT substrate specificity, establishing a foundation for the rational design of selective MBT domain inhibitors that may enable elucidation of their role in human biology and disease.
- Ontario Cancer Institute, Campbell Family Cancer Research Institute and Department of Medical Biophysics, University of Toronto, 101 College Street, Toronto, ON, Canada M5G 1L7.
Organizational Affiliation: