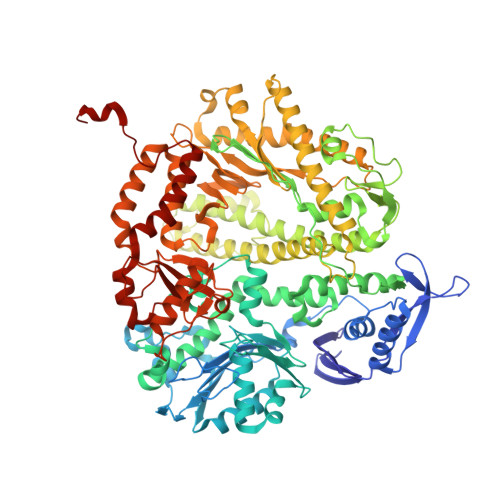
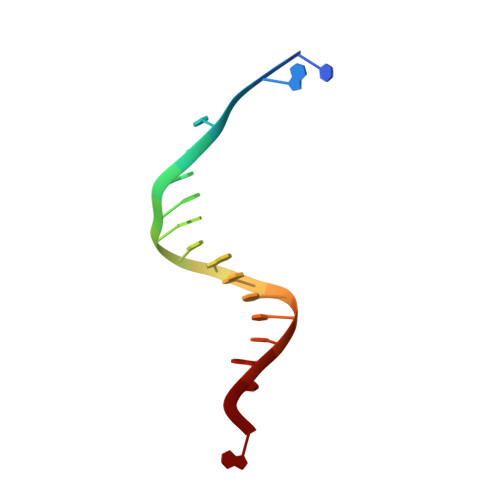
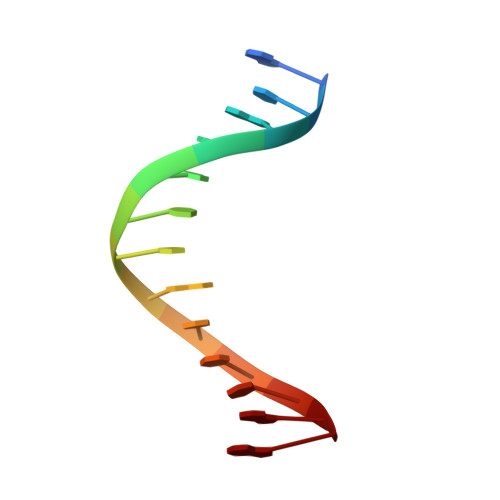
Contribution of Partial Charge Interactions and Base Stacking to the Efficiency of Primer Extension at and beyond Abasic Sites in DNA.
Xia, S., Vashishtha, A., Bulkley, D., Eom, S.H., Wang, J., Konigsberg, W.H.(2012) Biochemistry 51: 4922-4931
- PubMed: 22630605
- DOI: https://doi.org/10.1021/bi300296q
- Primary Citation of Related Structures:
4DTJ, 4DTM, 4DTN, 4DTO, 4DTP, 4DTR, 4DTS, 4DTU, 4DTX - PubMed Abstract:
During DNA synthesis, base stacking and Watson-Crick (WC) hydrogen bonding increase the stability of nascent base pairs when they are in a ternary complex. To evaluate the contribution of base stacking to the incorporation efficiency of dNTPs when a DNA polymerase encounters an abasic site, we varied the penultimate base pairs (PBs) adjacent to the abasic site using all 16 possible combinations. We then determined pre-steady-state kinetic parameters with an RB69 DNA polymerase variant and solved nine structures of the corresponding ternary complexes. The efficiency of incorporation for incoming dNTPs opposite an abasic site varied between 2- and 210-fold depending on the identity of the PB. We propose that the A rule can be extended to encompass the fact that DNA polymerase can bypass dA/abasic sites more efficiently than other dN/abasic sites. Crystal structures of the ternary complexes show that the surface of the incoming base was stacked against the PB's interface and that the kinetic parameters for dNMP incorporation were consistent with specific features of base stacking, such as surface area and partial charge-charge interactions between the incoming base and the PB. Without a templating nucleotide residue, an incoming dNTP has no base with which it can hydrogen bond and cannot be desolvated, so that these surrounding water molecules become ordered and remain on the PB's surface in the ternary complex. When these water molecules are on top of a hydrophobic patch on the PB, they destabilize the ternary complex, and the incorporation efficiency of incoming dNTPs is reduced.
- Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, Connecticut 06520-8114, USA.
Organizational Affiliation: