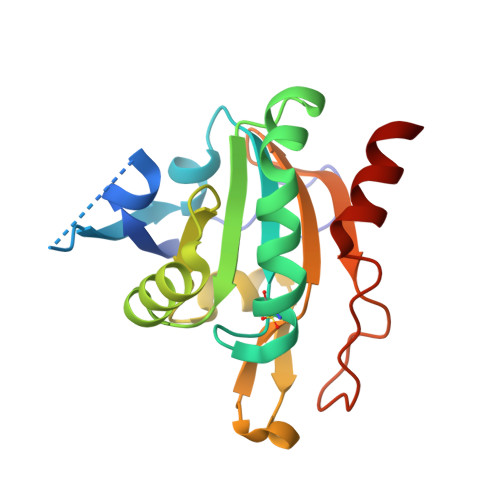
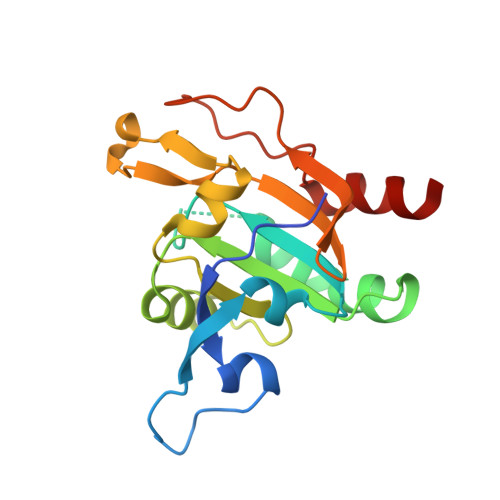
Plasmodium Falciparum Antioxidant Protein Reveals a Novel Mechanism for Balancing Turnover and Inactivation of Peroxiredoxins
Staudacher, V., Djuika, C.F., Koduka, J., Schlossarek, S., Kopp, J., Buechler, M., Lanzer, M., Deponte, M.(2015) Free Radic Biol Med 85: 228
- PubMed: 25952724
- DOI: https://doi.org/10.1016/j.freeradbiomed.2015.04.030
- Primary Citation of Related Structures:
4D73 - PubMed Abstract:
Life under aerobic conditions has shaped peroxiredoxins (Prx) as ubiquitous thiol-dependent hydroperoxidases and redox sensors. Structural features that balance the catalytically active or inactive redox states of Prx, and, therefore, their hydroperoxidase or sensor function, have so far been analyzed predominantly for Prx1-type enzymes. Here we identify and characterize two modulatory residues of the Prx5-type model enzyme PfAOP from the malaria parasite Plasmodium falciparum. Gain- and loss-of-function mutants reveal a correlation between the enzyme parameters and the inactivation susceptibility of PfAOP with the size of residue 109 and the presence or absence of a catalytically relevant but nonessential cysteine residue. Based on our kinetic data and the crystal structure of PfAOP(L109M), we suggest a novel mechanism for balancing the hydroperoxidase activity and inactivation susceptibility of Prx5-type enzymes. Our study provides unexpected insights into Prx structure-function relationships and contributes to our understanding of what makes Prx good enzymes or redox sensors.
- Department of Parasitology, Ruprecht-Karls University, D-69120 Heidelberg, Germany.
Organizational Affiliation: