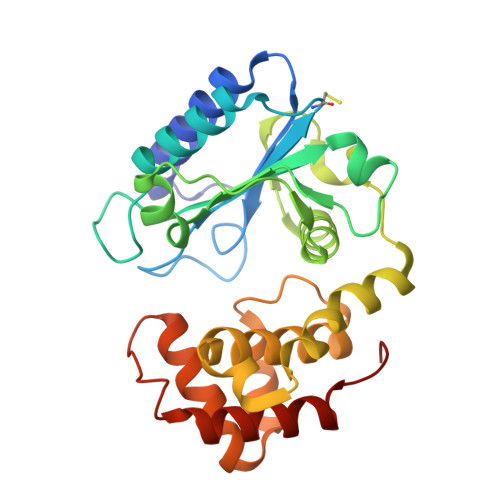
Structure of Aada from Salmonella Enterica: A Monomeric Aminoglycoside (3'')(9) Adenyltransferase.
Chen, Y., Nasvall, J., Wu, S., Andersson, D.I., Selmer, M.(2015) Acta Crystallogr D Biol Crystallogr 71: 2267
- PubMed: 26527143
- DOI: https://doi.org/10.1107/S1399004715016429
- Primary Citation of Related Structures:
4CS6 - PubMed Abstract:
Aminoglycoside resistance is commonly conferred by enzymatic modification of drugs by aminoglycoside-modifying enzymes such as aminoglycoside nucleotidyltransferases (ANTs). Here, the first crystal structure of an ANT(3'')(9) adenyltransferase, AadA from Salmonella enterica, is presented. AadA catalyses the magnesium-dependent transfer of adenosine monophosphate from ATP to the two chemically dissimilar drugs streptomycin and spectinomycin. The structure was solved using selenium SAD phasing and refined to 2.5 Å resolution. AadA consists of a nucleotidyltransferase domain and an α-helical bundle domain. AadA crystallizes as a monomer and is a monomer in solution as confirmed by small-angle X-ray scattering, in contrast to structurally similar homodimeric adenylating enzymes such as kanamycin nucleotidyltransferase. Isothermal titration calorimetry experiments show that ATP binding has to occur before binding of the aminoglycoside substrate, and structure analysis suggests that ATP binding repositions the two domains for aminoglycoside binding in the interdomain cleft. Candidate residues for ligand binding and catalysis were subjected to site-directed mutagenesis. In vivo resistance and in vitro binding assays support the role of Glu87 as the catalytic base in adenylation, while Arg192 and Lys205 are shown to be critical for ATP binding.
- Department of Cell and Molecular Biology, Uppsala University, Biomedical Center, Box 596, SE-751 24 Uppsala, Sweden.
Organizational Affiliation: