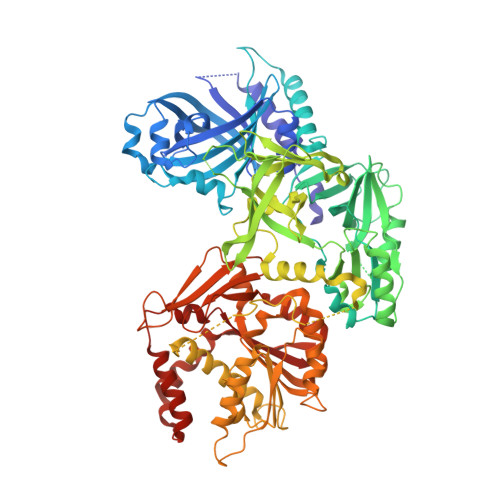
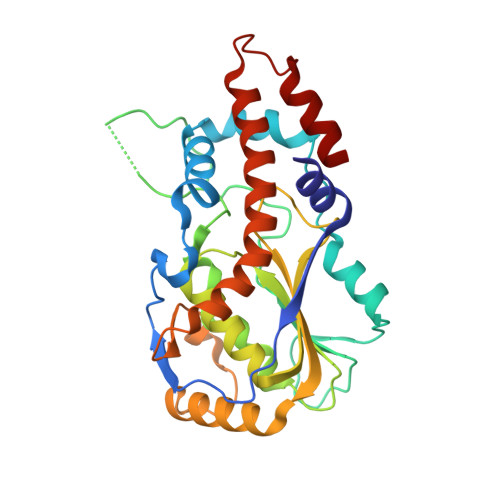
Crystal Structure of Vaccinia Virus Mrna Capping Enzyme Provides Insights Into the Mechanism and Evolution of the Capping Apparatus.
Kyrieleis, O.J.P., Chang, J., De La Pena, M., Shuman, S., Cusack, S.(2014) Structure 22: 452
- PubMed: 24607143
- DOI: https://doi.org/10.1016/j.str.2013.12.014
- Primary Citation of Related Structures:
4CKB, 4CKC, 4CKE - PubMed Abstract:
Vaccinia virus capping enzyme is a heterodimer of D1 (844 aa) and D12 (287 aa) polypeptides that executes all three steps in m(7)GpppRNA synthesis. The D1 subunit comprises an N-terminal RNA triphosphatase (TPase)-guanylyltransferase (GTase) module and a C-terminal guanine-N7-methyltransferase (MTase) module. The D12 subunit binds and allosterically stimulates the MTase module. Crystal structures of the complete D1⋅D12 heterodimer disclose the TPase and GTase as members of the triphosphate tunnel metalloenzyme and covalent nucleotidyltransferase superfamilies, respectively, albeit with distinctive active site features. An extensive TPase-GTase interface clamps the GTase nucleotidyltransferase and OB-fold domains in a closed conformation around GTP. Mutagenesis confirms the importance of the TPase-GTase interface for GTase activity. The D1⋅D12 structure complements and rationalizes four decades of biochemical studies of this enzyme, which was the first capping enzyme to be purified and characterized, and provides new insights into the origins of the capping systems of other large DNA viruses.
- European Molecular Biology Laboratory, Grenoble Outstation, 6 rue Horowitz, BP 181, 38042 Grenoble Cedex 9, France; University Grenoble Alpes-CNRS-EMBL Unit of Virus Host Cell Interactions, 6 rue Horowitz, BP 181, 38042 Grenoble Cedex 9, France.
Organizational Affiliation: