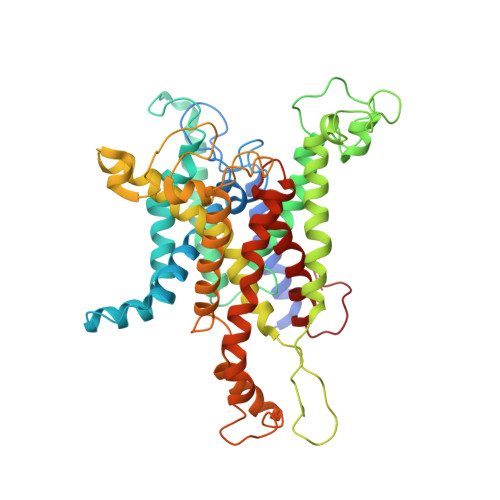
Structures of the Sec61 Complex Engaged in Nascent Peptide Translocation or Membrane Insertion.
Gogala, M., Becker, T., Beatrix, B., Armache, J., Barrio-Garcia, C., Berninghausen, O., Beckmann, R.(2014) Nature 506: 107
- PubMed: 24499919
- DOI: https://doi.org/10.1038/nature12950
- Primary Citation of Related Structures:
4CG5, 4CG6, 4CG7, 4V7E - PubMed Abstract:
The biogenesis of secretory as well as transmembrane proteins requires the activity of the universally conserved protein-conducting channel (PCC), the Sec61 complex (SecY complex in bacteria). In eukaryotic cells the PCC is located in the membrane of the endoplasmic reticulum where it can bind to translating ribosomes for co-translational protein transport. The Sec complex consists of three subunits (Sec61α, β and γ) and provides an aqueous environment for the translocation of hydrophilic peptides as well as a lateral opening in the Sec61α subunit that has been proposed to act as a gate for the membrane partitioning of hydrophobic domains. A plug helix and a so-called pore ring are believed to seal the PCC against ion flow and are proposed to rearrange for accommodation of translocating peptides. Several crystal and cryo-electron microscopy structures revealed different conformations of closed and partially open Sec61 and SecY complexes. However, in none of these samples has the translocation state been unambiguously defined biochemically. Here we present cryo-electron microscopy structures of ribosome-bound Sec61 complexes engaged in translocation or membrane insertion of nascent peptides. Our data show that a hydrophilic peptide can translocate through the Sec complex with an essentially closed lateral gate and an only slightly rearranged central channel. Membrane insertion of a hydrophobic domain seems to occur with the Sec complex opening the proposed lateral gate while rearranging the plug to maintain an ion permeability barrier. Taken together, we provide a structural model for the basic activities of the Sec61 complex as a protein-conducting channel.
- Gene Center and Center for integrated Protein Science Munich, Department of Biochemistry, Feodor-Lynen-Strasse 25, University of Munich, 81377 Munich, Germany.
Organizational Affiliation: