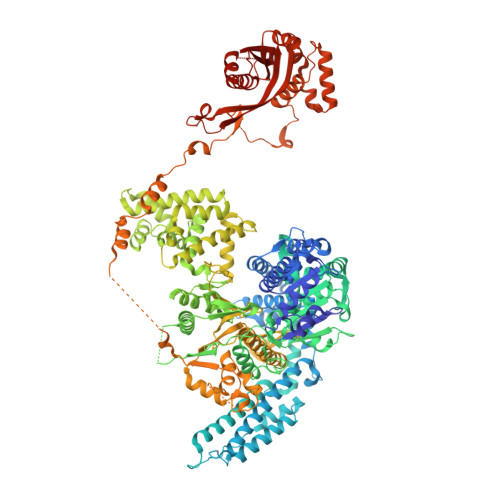
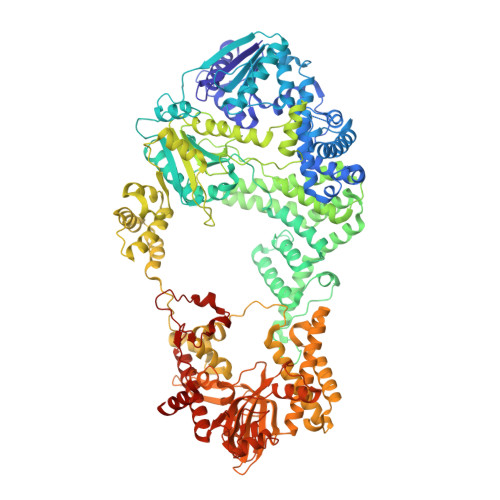
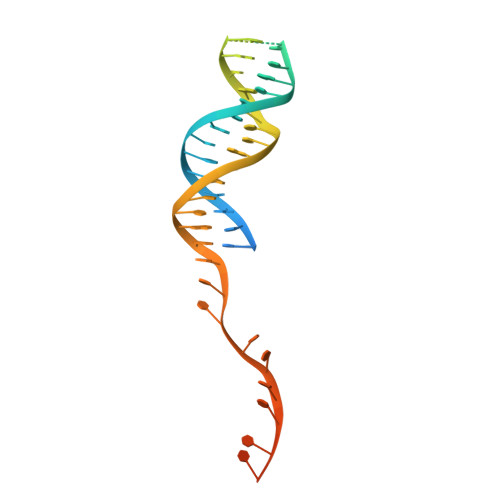
Structural Basis for Translocation by Addab Helicase-Nuclease and its Arrest at Chi Sites.
Krajewski, W.W., Fu, X., Wilkinson, M., Cronin, N.B., Dillingham, M.S., Wigley, D.B.(2014) Nature 508: 416
- PubMed: 24670664
- DOI: https://doi.org/10.1038/nature13037
- Primary Citation of Related Structures:
4CEH, 4CEI, 4CEJ - PubMed Abstract:
In bacterial cells, processing of double-stranded DNA breaks for repair by homologous recombination is dependent upon the recombination hotspot sequence χ (Chi) and is catalysed by either an AddAB- or RecBCD-type helicase-nuclease (reviewed in refs 3, 4). These enzyme complexes unwind and digest the DNA duplex from the broken end until they encounter a χ sequence, whereupon they produce a 3' single-stranded DNA tail onto which they initiate loading of the RecA protein. Consequently, regulation of the AddAB/RecBCD complex by χ is a key control point in DNA repair and other processes involving genetic recombination. Here we report crystal structures of Bacillus subtilis AddAB in complex with different χ-containing DNA substrates either with or without a non-hydrolysable ATP analogue. Comparison of these structures suggests a mechanism for DNA translocation and unwinding, suggests how the enzyme binds specifically to χ sequences, and explains how χ recognition leads to the arrest of AddAB (and RecBCD) translocation that is observed in single-molecule experiments.
- 1] Division of Structural Biology, Institute of Cancer Research, Chester Beatty Laboratories, 237 Fulham Road, London SW3 6JB, UK [2] CRT Discovery Laboratories, Department of Biological Sciences, Birkbeck, University of London, London WC1E 7HX, UK.
Organizational Affiliation: