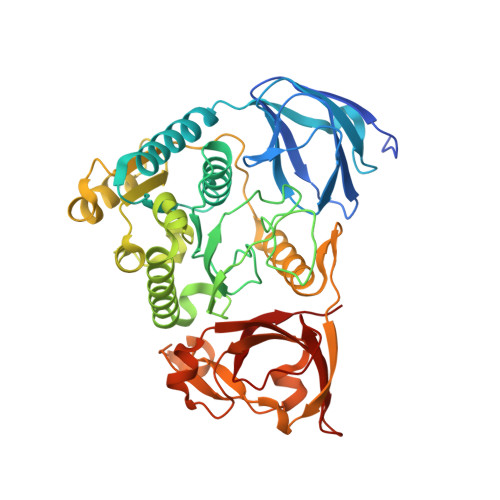
Structural basis for ATP loss by Clp1p in a G135R mutant protein.
Dupin, A.F., Fribourg, S.(2014) Biochimie 101: 203-207
- PubMed: 24508575
- DOI: https://doi.org/10.1016/j.biochi.2014.01.017
- Primary Citation of Related Structures:
4C0B, 4C0H - PubMed Abstract:
Pcf11p and Clp1p form a heterodimer and are subunits of the Cleavage Factor IA (CF IA), a complex that is involved in the maturation of the 3'-end of mRNAs in Saccharomyces cerevisiae. The role of Clp1p protein in polyadenylation remains elusive, as does the need for ATP binding by Clp1p. In order to obtain structural details at atomic resolution of point mutants of Clp1p, we solved the crystal structure of Clp1-1p (G135R) point mutant complexed with Pcf11p (454-563) domain. The Clp1-1p-Pcf11p structure provides the atomic details for ATP loss while the point mutation preserves intact the Pcf11p interaction surface of Clp1p. This provides a rationale for the absence of phenotype in the yeast clp1-1 strain. Additionally, the structure allows for the description of an extended binding interface of Pcf11p with Clp1p which is likely to be S. cerevisiae specific.
- Univ. Bordeaux, IECB, F-33607 Pessac, France; INSERM, U869, F-33077 Pessac, France.
Organizational Affiliation: