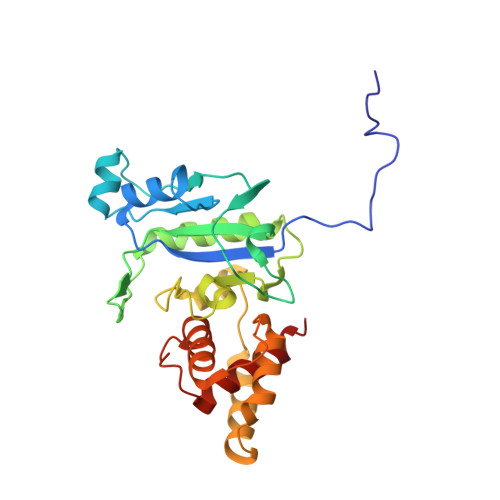
Cell-Wall Remodeling by the Zinc-Protease Ampdh3 from Pseudomonas Aeruginosa.
Lee, M., Artola-Recolons, C., Carrasco-Lopez, C., Martinez-Caballero, S., Hesek, D., Spink, E., Lastochkin, E., Zhang, W., Hellman, L.M., Boggess, B., Hermoso, J.A., Mobashery, S.(2013) J Am Chem Soc 135: 12604
- PubMed: 23931161
- DOI: https://doi.org/10.1021/ja407445x
- Primary Citation of Related Structures:
4BXD, 4BXE, 4BXJ - PubMed Abstract:
Bacterial cell wall is a polymer of considerable complexity that is in constant equilibrium between synthesis and recycling. AmpDh3 is a periplasmic zinc protease of Pseudomonas aeruginosa , which is intimately involved in cell-wall remodeling. We document the hydrolytic reactions that this enzyme performs on the cell wall. The process removes the peptide stems from the peptidoglycan, the major constituent of the cell wall. We document that the majority of the reactions of this enzyme takes place on the polymeric insoluble portion of the cell wall, as opposed to the fraction that is released from it. We show that AmpDh3 is tetrameric both in crystals and in solution. Based on the X-ray structures of the enzyme in complex with two synthetic cell-wall-based ligands, we present for the first time a model for a multivalent anchoring of AmpDh3 onto the cell wall, which lends itself to its processive remodeling.
- Department of Chemistry and Biochemistry, University of Notre Dame, Notre Dame, Indiana 46556, USA.
Organizational Affiliation: