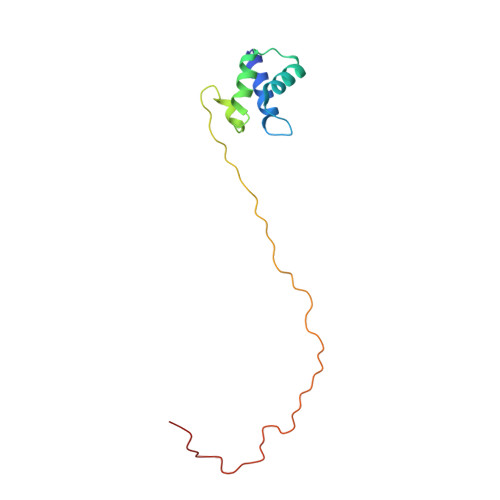
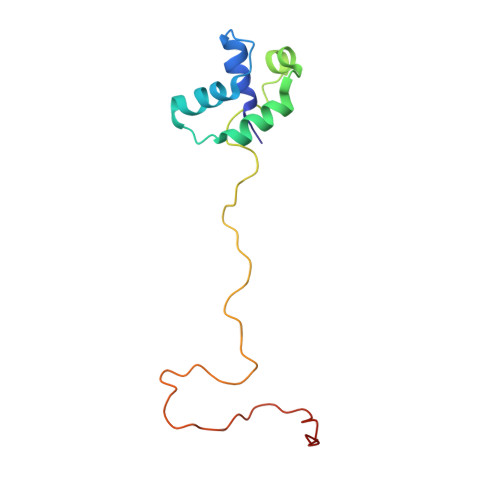
Solution Structure of Human P1P2 Heterodimer Provides Insights Into the Role of Eukaryotic Stalk in Recruiting the Ribosome-Inactivating Protein Trichosanthin to the Ribosome.
Lee, K.M., Yusa, K., Chu, L.O., Wing-Heng Yu, C., Oono, M., Miyoshi, T., Ito, K., Shaw, P.C., Wong, K.B., Uchiumi, T.(2013) Nucleic Acids Res 41: 8776
- PubMed: 23892290
- DOI: https://doi.org/10.1093/nar/gkt636
- Primary Citation of Related Structures:
4BEH - PubMed Abstract:
Lateral ribosomal stalk is responsible for binding and recruiting translation factors during protein synthesis. The eukaryotic stalk consists of one P0 protein with two copies of P1•P2 heterodimers to form a P0(P1•P2)₂ pentameric P-complex. Here, we have solved the structure of full-length P1•P2 by nuclear magnetic resonance spectroscopy. P1 and P2 dimerize via their helical N-terminal domains, whereas the C-terminal tails of P1•P2 are unstructured and can extend up to ∼125 Å away from the dimerization domains. (15)N relaxation study reveals that the C-terminal tails are flexible, having a much faster internal mobility than the N-terminal domains. Replacement of prokaryotic L10(L7/L12)₄/L11 by eukaryotic P0(P1•P2)₂/eL12 rendered Escherichia coli ribosome, which is insensitive to trichosanthin (TCS), susceptible to depurination by TCS and the C-terminal tail was found to be responsible for this depurination. Truncation and insertion studies showed that depurination of hybrid ribosome is dependent on the length of the proline-alanine rich hinge region within the C-terminal tail. All together, we propose a model that recruitment of TCS to the sarcin-ricin loop required the flexible C-terminal tail, and the proline-alanine rich hinge region lengthens this C-terminal tail, allowing the tail to sweep around the ribosome to recruit TCS.
- School of Life Sciences, Centre for Protein Science and Crystallography, The Chinese University of Hong Kong, Shatin, Hong Kong, China and Department of Biology, Faculty of Science, Niigata University, Ikarashi 2-8050, Nishi-ku, Niigata 950-2181, Japan.
Organizational Affiliation: