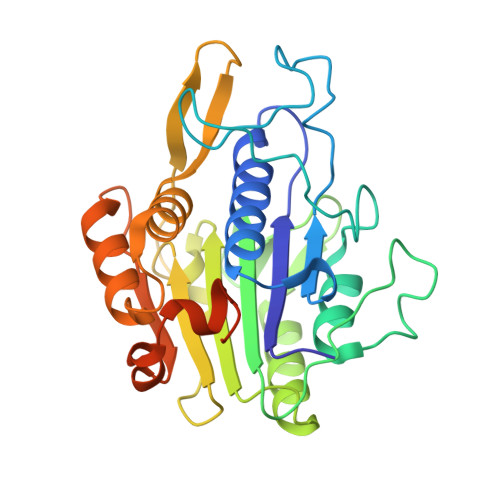
Mechanistic and Structural Studies on Legumain Explain its Zymogenicity, Distinct Activation Pathways, and Regulation
Dall, E., Brandstetter, H.(2013) Proc Natl Acad Sci U S A 110: 10940
- PubMed: 23776206
- DOI: https://doi.org/10.1073/pnas.1300686110
- Primary Citation of Related Structures:
4AW9, 4AWA, 4AWB, 4FGU - PubMed Abstract:
The cysteine protease legumain plays important functions in immunity and cancer at different cellular locations, some of which appeared conflicting with its proteolytic activity and stability. Here, we report crystal structures of legumain in the zymogenic and fully activated form in complex with different substrate analogs. We show that the eponymous asparagine-specific endopeptidase activity is electrostatically generated by pH shift. Completely unexpectedly, the structure points toward a hidden carboxypeptidase activity that develops upon proteolytic activation with the release of an activation peptide. These activation routes reconcile the enigmatic pH stability of legumain, e.g., lysosomal, nuclear, and extracellular activities with relevance in immunology and cancer. Substrate access and turnover is controlled by selective protonation of the S1 pocket (KM) and the catalytic nucleophile (kcat), respectively. The multibranched and context-dependent activation process of legumain illustrates how proteases can act not only as signal transducers but as decision makers.
- Department of Molecular Biology, University of Salzburg, A-5020 Salzburg, Austria.
Organizational Affiliation: