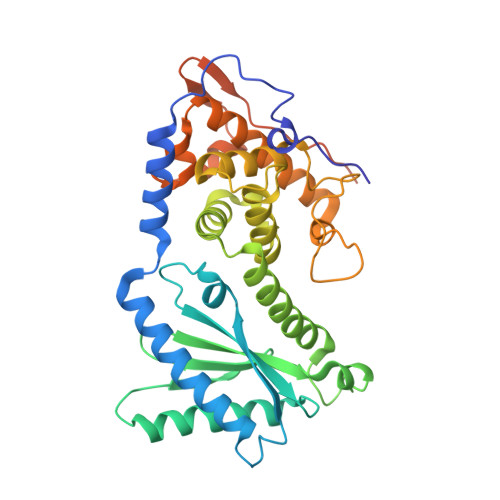
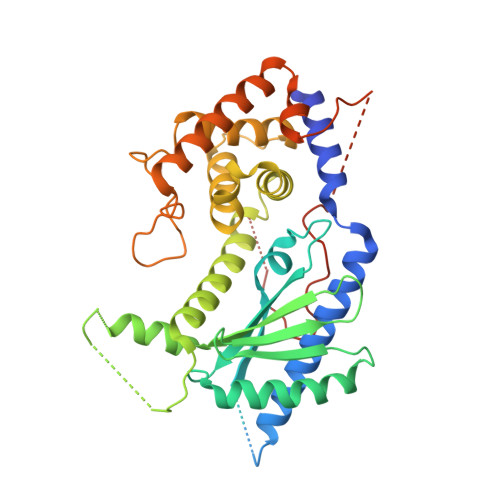
NF45 Dimerizes with NF90, Zfr and Spnr Via a Conserved Domain that Has a Nucleotidyltransferase Fold.
Wolkowicz, U.M., Cook, A.G.(2012) Nucleic Acids Res 40: 9356
- PubMed: 22833610
- DOI: https://doi.org/10.1093/nar/gks696
- Primary Citation of Related Structures:
4AT7, 4AT8, 4AT9, 4ATB - PubMed Abstract:
Nuclear factors NF90 and NF45 form a complex involved in a variety of cellular processes and are thought to affect gene expression both at the transcriptional and translational level. In addition, this complex affects the replication of several viruses through direct interactions with viral RNA. NF90 and NF45 dimerize through their common 'DZF' domain (domain associated with zinc fingers). NF90 has additional double-stranded RNA-binding domains that likely mediate its association with target RNAs. We present the crystal structure of the NF90/NF45 dimerization complex at 1.9-Å resolution. The DZF domain shows structural similarity to the template-free nucleotidyltransferase family of RNA modifying enzymes. However, both NF90 and NF45 have lost critical catalytic residues during evolution and are therefore not functional enzymes. Residues on NF90 that make up its interface with NF45 are conserved in two related proteins, spermatid perinuclear RNA-binding protein (SPNR) and zinc-finger RNA-binding protein (Zfr). Using a co-immunoprecipitation assay and site-specific mutants, we demonstrate that NF45 is also able to recognize SPNR and Zfr through the same binding interface, revealing that NF45 is able to form a variety of cellular complexes with other DZF-domain proteins.
- Wellcome Trust Centre for Cell Biology, University of Edinburgh, Michael Swann Building, Edinburgh, Midlothian EH9 3JR, UK.
Organizational Affiliation: