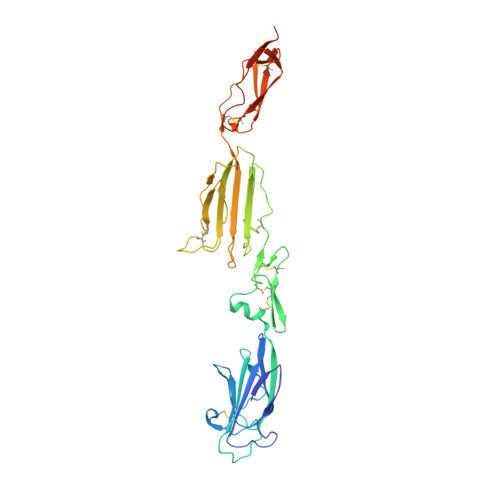
Crystal Structure and Functional Characterization of the Complement Regulator Mannose-Binding Lectin (Mbl)/Ficolin-Associated Protein-1 (Map-1).
Skjoedt, M.O., Roversi, P., Hummelshoj, T., Palarasah, Y., Rosbjerg, A., Johnson, S., Lea, S.M., Garred, P.(2012) J Biological Chem 287: 32913
- PubMed: 22854970
- DOI: https://doi.org/10.1074/jbc.M112.386680
- Primary Citation of Related Structures:
4AQB - PubMed Abstract:
The human lectin complement pathway activation molecules comprise mannose-binding lectin (MBL) and ficolin-1, -2, and -3 in complex with associated serine proteases MASP-1, -2, and -3 and the non-enzymatic small MBL associated protein or sMAP. Recently, a novel plasma protein named MBL/ficolin-associated protein-1 (MAP-1) was identified in humans. This protein is the result of a differential splicing of the MASP1 gene and includes the major part of the heavy chain but lacks the serine protease domain. We investigated the direct interactions of MAP-1 and MASP-3 with ficolin-3 and MBL using surface plasmon resonance and found affinities around 5 nm and 2.5 nm, respectively. We studied structural aspects of MAP-1 and could show by multi-angle laser light scattering that MAP-1 forms a calcium-dependent homodimer in solution. We were able to determine the crystal structure of MAP-1, which also contains a head-to-tail dimer ∼146 Å long. This structure of MAP-1 also enables modeling and assembly of the MASP-1 molecule in its entirety. Finally we found that MAP-1 competes with all three MASPs for ligand binding and is able to mediate a strong dose-dependent inhibitory effect on the lectin pathway activation, as measured by levels of C3 and C9.
- Laboratory of Molecular Medicine, Department of Clinical Immunology, Rigshospitalet, Faculty of Health Sciences, University of Copenhagen, DK 2100 Copenhagen, Denmark. moskjoedt@gmail.com
Organizational Affiliation: