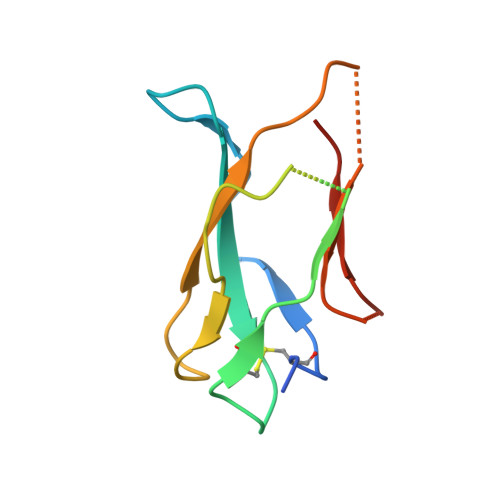
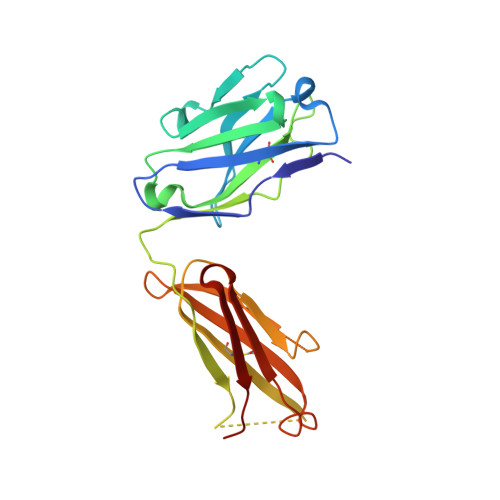
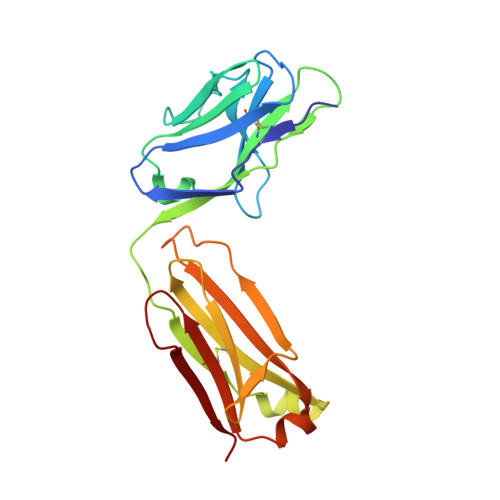
Structural Analysis of a Dengue Cross-Reactive Antibody Complexed with Envelope Domain III Reveals the Molecular Basis of Cross-Reactivity.
Midgley, C.M., Flanagan, A., Tran, H.B., Dejnirattisai, W., Chawansuntati, K., Jumnainsong, A., Wongwiwat, W., Duangchinda, T., Mongkolsapaya, J., Grimes, J.M., Screaton, G.R.(2012) J Immunol 188: 4971
- PubMed: 22491255
- DOI: https://doi.org/10.4049/jimmunol.1200227
- Primary Citation of Related Structures:
4AL8, 4ALA, 4AM0 - PubMed Abstract:
Dengue virus infections are still increasing at an alarming rate in tropical and subtropical countries, underlying the need for a dengue vaccine. Although it is relatively easy to generate Ab responses to dengue virus, low avidity or low concentrations of Ab may enhance infection of FcR-bearing cells with clinical impact, posing a challenge to vaccine production. In this article, we report the characterization of a mAb, 2H12, which is cross-reactive to all four serotypes in the dengue virus group. Crystal structures of 2H12-Fab in complex with domain III of the envelope protein from three dengue serotypes have been determined. 2H12 binds to the highly conserved AB loop of domain III of the envelope protein that is poorly accessible in the mature virion. 2H12 neutralization varied between dengue serotypes and strains; in particular, dengue serotype 2 was not neutralized. Because the 2H12-binding epitope was conserved, this variation in neutralization highlights differences between dengue serotypes and suggests that significant conformational changes in the virus must take place for Ab binding. Surprisingly, 2H12 facilitated little or no enhancement of infection. These data provide a structural basis for understanding Ab neutralization and enhancement of infection, which is crucial for the development of future dengue vaccines.
- Department of Medicine, Hammersmith Hospital Campus, Imperial College London, London W12 0NN, United Kingdom.
Organizational Affiliation: