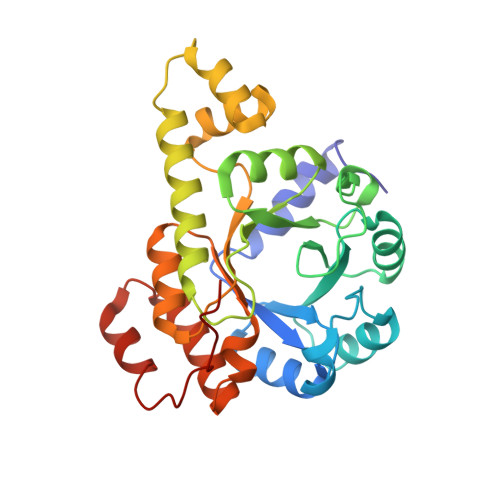
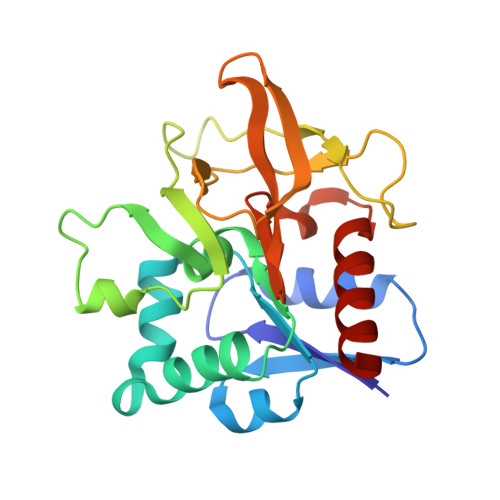
Assembly of the Eukaryotic Plp-Synthase Complex from Plasmodium and Activation of the Pdx1 Enzyme.
Guedez, G., Hipp, K., Windeisen, V., Derrer, B., Gengenbacher, M., Boettcher, B., Sinning, I., Kappes, B., Tews, I.(2012) Structure 20: 172
- PubMed: 22244765
- DOI: https://doi.org/10.1016/j.str.2011.11.015
- Primary Citation of Related Structures:
4ADS, 4ADT, 4ADU - PubMed Abstract:
Biosynthesis of vitamins is fundamental to malaria parasites. Plasmodia synthesize the active form of vitamin B(6) (pyridoxal 5'-phosphate, PLP) using a PLP synthase complex. The EM analysis shown here reveals a random association pattern of up to 12 Pdx2 glutaminase subunits to the dodecameric Pdx1 core complex. Interestingly, Plasmodium falciparum PLP synthase organizes in fibers. The crystal structure shows differences in complex formation to bacterial orthologs as interface variations. Alternative positioning of an α helix distinguishes an open conformation from a closed state when the enzyme binds substrate. The pentose substrate is covalently attached through its C1 and forms a Schiff base with Lys84. Ammonia transfer between Pdx2 glutaminase and Pdx1 active sites is regulated by a transient tunnel. The mutagenesis analysis allows defining the requirement for conservation of critical methionines, whereas there is also plasticity in ammonia tunnel construction as seen from comparison across different species.
- Heidelberg University Biochemistry Center (BZH), Im Neuenheimer Feld 328, 69120 Heidelberg, Germany.
Organizational Affiliation: