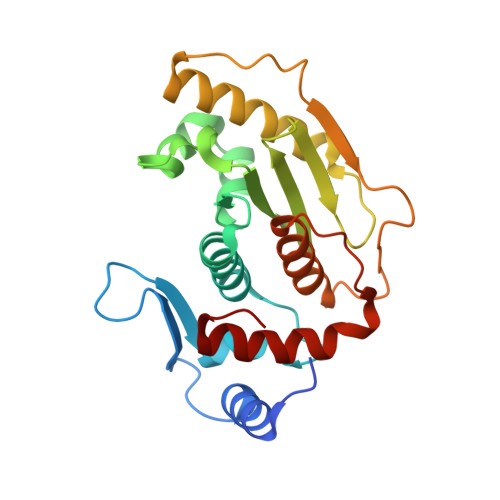
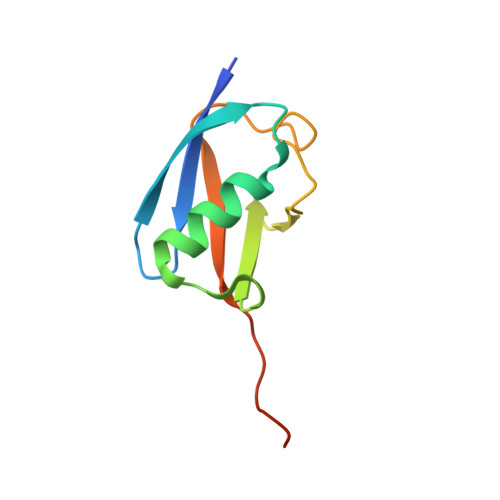
Structural Insights Into the Senp6 Loop1 Structure in Complex with Sumo2.
Alegre, K.O., Reverter, D.(2014) Protein Sci 23: 433
- PubMed: 24424631
- DOI: https://doi.org/10.1002/pro.2425
- Primary Citation of Related Structures:
3ZO5 - PubMed Abstract:
The SENP proteases regulate the SUMO conjugates in the cell by cleaving SUMO from target proteins. SENP6 and SENP7 are the most divergent members of the SENP/ULP protease family in humans by the presence of insertions in their catalytic domains. Loop1 insertion is determinant for the SUMO2/3 activity and specificity on SENP6 and SENP7. To gain structural insights into the role of Loop1, we have designed a chimeric SENP2 with the insertion of Loop1 into its sequence. The structure of SENP2-Loop1 in complex with SUMO2 was solved at 2.15 Å resolution, and reveals the details of an interface exclusive to SENP6/7 and the formation of unique contacts between both proteins. Interestingly, functional data with SUMO substrates showed an increase of the proteolytic activity in the SENP2-Loop1 chimera for diSUMO2 and polySUMO2 substrates.
- Institut de Biotecnologia i de Biomedicina and Departament de Bioquímica i Biologia Molecular, Universitat Autònoma de Barcelona, 08193, Bellaterra, Spain.
Organizational Affiliation: