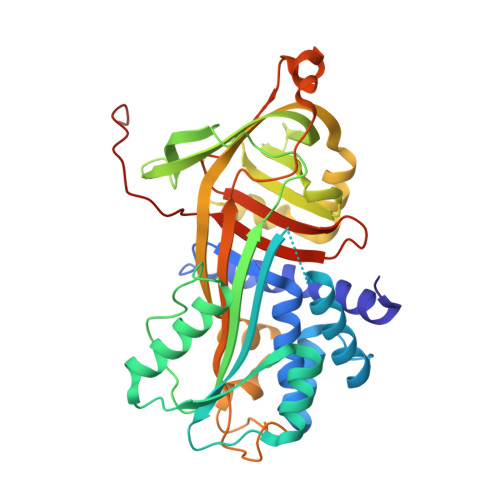
Molecular Basis for the Action of the Collagen-Specific Chaperone Hsp47/Serpinh1 and its Structure-Specific Client Recognition.
Widmer, C., Gebauer, J.M., Brunstein, E., Rosenbaum, S., Zaucke, F., Drogemuller, C., Leeb, T., Baumann, U.(2012) Proc Natl Acad Sci U S A 109: 13243
- PubMed: 22847422
- DOI: https://doi.org/10.1073/pnas.1208072109
- Primary Citation of Related Structures:
3ZHA, 4AU2, 4AU3, 4AU4, 4AXY - PubMed Abstract:
Collagen is the most abundant protein in animals and is a major component of the extracellular matrix in tissues such as skin and bone. A distinctive structural feature of all collagen types is a unique triple-helical structure formed by tandem repeats of the consensus sequence Xaa-Yaa-Gly, in which Xaa and Yaa frequently are proline and hydroxyproline, respectively. Hsp47/SERPINH1 is a procollagen-specific molecular chaperone that, unlike other chaperones, specifically recognizes the folded conformation of its client. Reduced functional levels of Hsp47 were reported in severe recessive forms of osteogenesis imperfecta, and homozygous knockout is lethal in mice. Here we present crystal structures of Hsp47 in its free form and in complex with homotrimeric synthetic collagen model peptides, each comprising one Hsp47-binding site represented by an arginine at the Yaa-position of a Xaa-Yaa-Gly triplet. Two of these three binding sites in the triple helix are occupied by Hsp47 molecules, which bind in a head-to-head fashion, thus making extensive contacts with the leading and trailing strands of the collagen triple helix. The important arginine residue within the Xaa-Arg-Gly triplet is recognized by a conserved aspartic acid. The structures explain the stabilization of the triple helix as well as the inhibition of collagen-bundle formation by Hsp47. In addition, we propose a pH-dependent substrate release mechanism based on a cluster of histidine residues.
- Department of Chemistry and Biochemistry, Vetsuisse Faculty University of Bern, CH-3012 Bern, Switzerland.
Organizational Affiliation: