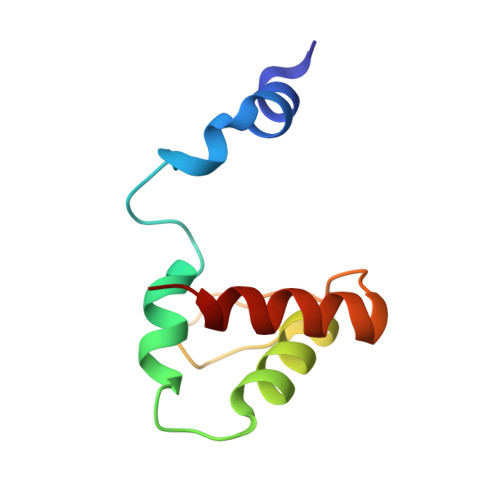
Domain-Swapped Dimer of Pseudomonas aeruginosa Cytochrome c551: Structural Insights into Domain Swapping of Cytochrome c Family Proteins
Nagao, S., Ueda, M., Osuka, H., Komori, H., Kamikubo, H., Kataoka, M., Higuchi, Y., Hirota, S.(2015) PLoS One 10: e0123653-e0123653
- PubMed: 25853415
- DOI: https://doi.org/10.1371/journal.pone.0123653
- Primary Citation of Related Structures:
3X39 - PubMed Abstract:
Cytochrome c (cyt c) family proteins, such as horse cyt c, Pseudomonas aeruginosa cytochrome c551 (PA cyt c551), and Hydrogenobacter thermophilus cytochrome c552 (HT cyt c552), have been used as model proteins to study the relationship between the protein structure and folding process. We have shown in the past that horse cyt c forms oligomers by domain swapping its C-terminal helix, perturbing the Met-heme coordination significantly compared to the monomer. HT cyt c552 forms dimers by domain swapping the region containing the N-terminal α-helix and heme, where the heme axial His and Met ligands belong to different protomers. Herein, we show that PA cyt c551 also forms domain-swapped dimers by swapping the region containing the N-terminal α-helix and heme. The secondary structures of the M61A mutant of PA cyt c551 were perturbed slightly and its oligomer formation ability decreased compared to that of the wild-type protein, showing that the stability of the protein secondary structures is important for domain swapping. The hinge loop of domain swapping for cyt c family proteins corresponded to the unstable region specified by hydrogen exchange NMR measurements for the monomer, although the swapping region differed among proteins. These results show that the unstable loop region has a tendency to become a hinge loop in domain-swapped proteins.
- Graduate School of Materials Science, Nara Institute of Science and Technology, 8916-5 Takayama, Ikoma, Nara 630-0192, Japan.
Organizational Affiliation: