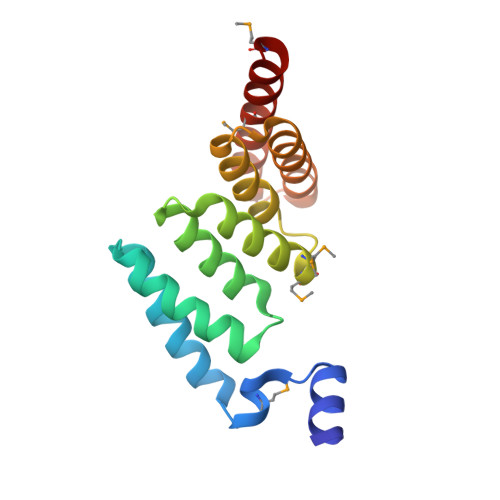
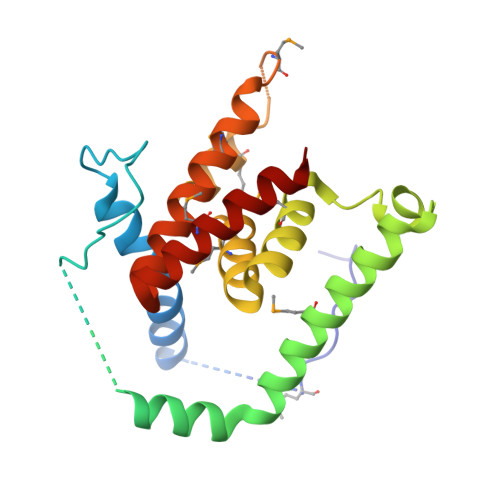
Structure of AcrH-AopB Chaperone-Translocator Complex Reveals a Role for Membrane Hairpins in Type III Secretion System Translocon Assembly
Nguyen, V.S., Jobichen, C., Tan, K.W., Tan, Y.W., Chan, S.L., Ramesh, K., Yuan, Y., Hong, Y., Seetharaman, J., Leung, K.Y., Sivaraman, J., Mok, Y.K.(2015) Structure 23: 2022-2031
- PubMed: 26439768
- DOI: https://doi.org/10.1016/j.str.2015.08.014
- Primary Citation of Related Structures:
3WXX - PubMed Abstract:
Type III secretion systems (T3SSs) are adopted by pathogenic bacteria for the transport of effector proteins into host cells through the translocon pore composed of major and minor translocator proteins. Both translocators require a dedicated chaperone for solubility. Despite tremendous efforts in the past, structural information regarding the chaperone-translocator complex and the topology of the translocon pore have remained elusive. Here, we report the crystal structure of the major translocator, AopB, from Aeromonas hydrophila AH-1 in complex with its chaperone, AcrH. Overall, the structure revealed unique interactions between the various interfaces of AopB and AcrH, with the N-terminal "molecular anchor" of AopB crossing into the "N-terminal arm" of AcrH. AopB adopts a novel fold, and its transmembrane regions form two pairs of helical hairpins. From these structural studies and associated cellular assays, we deduced the topology of the assembled T3SS translocon; both termini remain extracellular after membrane insertion.
- Department of Biological Sciences, 14 Science Drive 4, National University of Singapore, 117543 Singapore.
Organizational Affiliation: