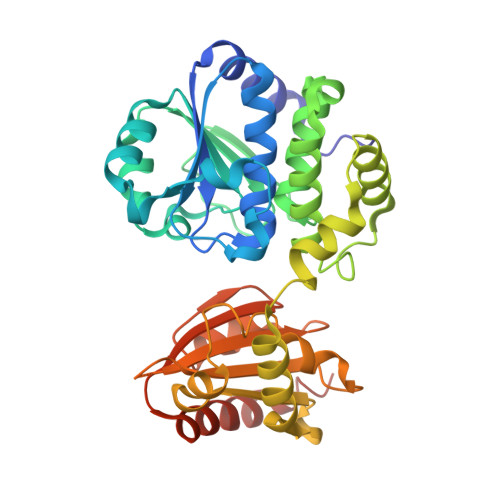
Crystal structure of the ligand-binding form of nanoRNase from Bacteroides fragilis, a member of the DHH/DHHA1 phosphoesterase family of proteins.
Uemura, Y., Nakagawa, N., Wakamatsu, T., Kim, K., Montelione, G.T., Hunt, J.F., Kuramitsu, S., Masui, R.(2013) FEBS Lett 587: 2669-2674
- PubMed: 23851074
- DOI: https://doi.org/10.1016/j.febslet.2013.06.053
- Primary Citation of Related Structures:
3W5W - PubMed Abstract:
NanoRNase (Nrn) specifically degrades nucleoside 3',5'-bisphosphate and the very short RNA, nanoRNA, during the final step of mRNA degradation. The crystal structure of Nrn in complex with a reaction product GMP was determined. The overall structure consists of two domains that are interconnected by a flexible loop and form a cleft. Two Mn²⁺ ions are coordinated by conserved residues in the DHH motif of the N-terminal domain. GMP binds near the DHHA1 motif region in the C-terminal domain. Our structure enables us to predict the substrate-bound form of Nrn as well as other DHH/DHHA1 phosphoesterase family proteins.
- Graduate School of Frontier Biosciences, Osaka University, Suita, Osaka 565-0871, Japan.
Organizational Affiliation: