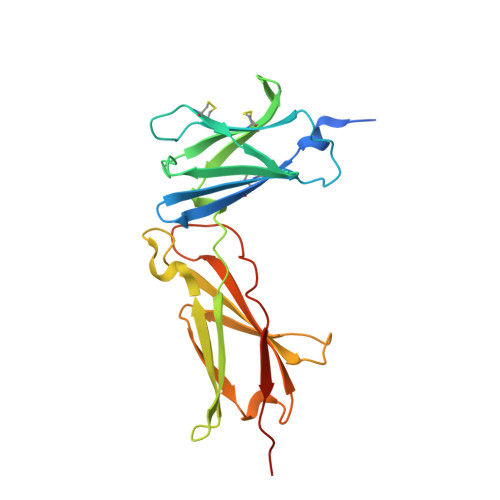
Structural reorganization of the interleukin-7 signaling complex.
McElroy, C.A., Holland, P.J., Zhao, P., Lim, J.M., Wells, L., Eisenstein, E., Walsh, S.T.(2012) Proc Natl Acad Sci U S A 109: 2503-2508
- PubMed: 22308406
- DOI: https://doi.org/10.1073/pnas.1116582109
- Primary Citation of Related Structures:
3UP1 - PubMed Abstract:
We report here an unliganded receptor structure in the common gamma-chain (γ(c)) family of receptors and cytokines. The crystal structure of the unliganded form of the interleukin-7 alpha receptor (IL-7Rα) extracellular domain (ECD) at 2.15 Å resolution reveals a homodimer forming an "X" geometry looking down onto the cell surface with the C termini of the two chains separated by 110 Å and the dimer interface comprising residues critical for IL-7 binding. Further biophysical studies indicate a weak association of the IL-7Rα ECDs but a stronger association between the γ(c)/IL-7Rα ECDs, similar to previous studies of the full-length receptors on CD4(+) T cells. Based on these and previous results, we propose a molecular mechanism detailing the progression from the inactive IL-7Rα homodimer and IL-7Rα-γ(c) heterodimer to the active IL-7-IL-7Rα-γ(c) ternary complex whereby the two receptors undergo at least a 90° rotation away from the cell surface, moving the C termini of IL-7Rα and γ(c) from a distance of 110 Å to less than 30 Å at the cell surface. This molecular mechanism can be used to explain recently discovered IL-7- and γ(c)-independent gain-of-function mutations in IL-7Rα from B- and T-cell acute lymphoblastic leukemia patients. The mechanism may also be applicable to other γ(c) receptors that form inactive homodimers and heterodimers independent of their cytokines.
- Battelle Biomedical Research Center, Columbus, OH 43201, USA.
Organizational Affiliation: